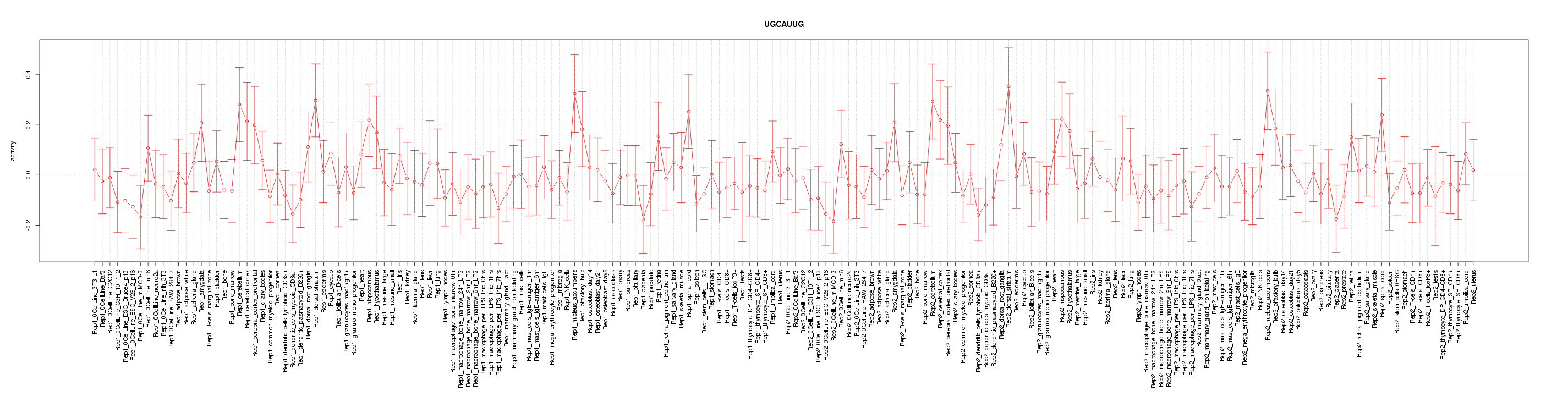
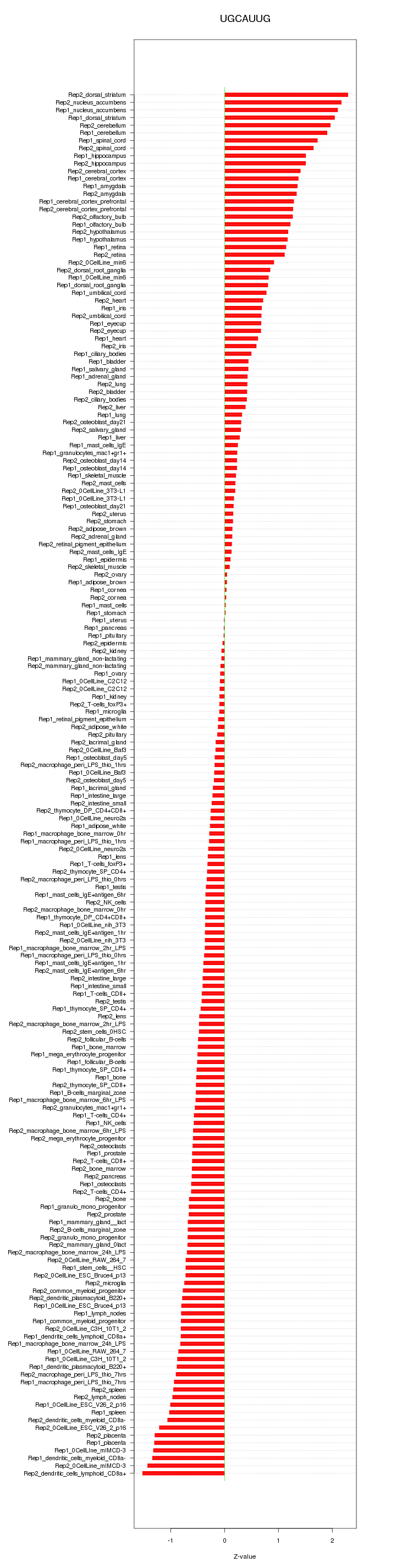
|
chr2_+_164793487
|
23.094
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr17_+_25945986
|
13.839
|
NM_001164225
|
Fbxl16
|
F-box and leucine-rich repeat protein 16
|
|
chr3_-_84024766
|
13.461
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr15_+_100701068
|
13.325
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr15_+_100766701
|
12.863
|
NM_011323
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr5_+_130845319
|
12.300
|
NM_181045
|
Caln1
|
calneuron 1
|
|
chr5_+_130924661
|
10.938
|
NM_021371
|
Caln1
|
calneuron 1
|
|
chr13_-_105844882
|
10.923
|
NM_029879
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr6_+_71657791
|
10.379
|
NM_178608
|
Reep1
|
receptor accessory protein 1
|
|
chr1_-_25886551
|
9.926
|
NM_175642
|
Bai3
|
brain-specific angiogenesis inhibitor 3
|
|
chr6_-_92517359
|
9.386
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_92656148
|
9.176
|
NM_001081146
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr7_-_148615247
|
8.618
|
NM_021316
|
Cend1
|
cell cycle exit and neuronal differentiation 1
|
|
chr6_-_92484838
|
8.535
|
NM_001134459
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr11_-_103815361
|
8.263
|
NM_008740
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr9_-_56533004
|
8.262
|
NM_181074
|
Lingo1
|
leucine rich repeat and Ig domain containing 1
|
|
chr17_-_64681166
|
8.227
|
NM_001025309
NM_144859
|
Pja2
|
praja 2, RING-H2 motif containing
|
|
chr6_-_92431385
|
8.149
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr19_-_58935470
|
7.879
|
NM_175199
|
Hspa12a
|
heat shock protein 12A
|
|
chr3_-_81949667
|
7.288
|
NM_021896
|
Gucy1a3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr7_+_94732677
|
7.196
|
NM_001081414
NM_001143834
|
Grm5
|
glutamate receptor, metabotropic 5
|
|
chr1_-_77511653
|
7.148
|
NM_007936
|
Epha4
|
Eph receptor A4
|
|
chr9_-_54349409
|
6.941
|
NM_172771
|
Dmxl2
|
Dmx-like 2
|
|
chr2_+_138104219
|
6.877
|
NM_145534
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr2_+_138082301
|
6.797
|
NM_001025431
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr3_-_26052228
|
6.751
|
NM_001163387
|
Nlgn1
|
neuroligin 1
|
|
chr12_-_14158843
|
6.706
|
NM_029007
|
Fam84a
|
family with sequence similarity 84, member A
|
|
chr3_+_64913564
|
6.685
|
NM_010597
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr3_-_26230791
|
6.669
|
NM_138666
|
Nlgn1
|
neuroligin 1
|
|
chr10_+_4424140
|
6.520
|
NM_001161822
NM_019958
|
Rgs17
|
regulator of G-protein signaling 17
|
|
chr2_+_156246644
|
6.240
|
NM_001003815
NM_013510
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr16_-_67621054
|
6.110
|
NM_001145977
NM_178721
|
Cadm2
|
cell adhesion molecule 2
|
|
chr10_-_49508472
|
6.066
|
NM_001111268
NM_010349
|
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2)
|
|
chr13_+_118391126
|
6.012
|
NM_010408
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1
|
|
chr19_+_38339167
|
5.887
|
NM_020278
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr6_+_4853319
|
5.880
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr13_+_109444370
|
5.824
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr6_-_28781687
|
5.451
|
NM_138682
|
Lrrc4
|
leucine rich repeat containing 4
|
|
chr5_-_71233855
|
5.038
|
NM_010252
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1
|
|
chr12_+_25516433
|
4.758
|
NM_001083341
NM_026037
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr3_+_5218545
|
4.628
|
NM_030708
|
Zfhx4
|
zinc finger homeodomain 4
|
|
chr2_+_156301543
|
4.547
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr6_-_119058206
|
4.526
|
NM_001159533
NM_001159534
NM_001159535
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr6_-_119146426
|
4.419
|
NM_009781
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr18_+_57292743
|
4.382
|
NM_001001979
|
Megf10
|
multiple EGF-like-domains 10
|
|
chr2_+_35477498
|
4.354
|
NM_001114124
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr2_+_35547418
|
3.977
|
NM_001001602
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chrX_+_20265590
|
3.706
|
NM_011049
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr7_+_129432585
|
3.528
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr2_+_76244578
|
3.349
|
NM_145525
|
Osbpl6
|
oxysterol binding protein-like 6
|
|
chr14_-_31304341
|
3.327
|
NM_028981
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr2_-_32306938
|
3.263
|
NM_001164357
NM_146118
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25
|
|
chr2_-_134469806
|
3.199
|
NM_029148
|
Tmx4
|
thioredoxin-related transmembrane protein 4
|
|
chr4_-_8166182
|
3.195
|
NM_007592
|
Car8
|
carbonic anhydrase 8
|
|
chr6_-_149050201
|
3.140
|
NM_177192
|
Dennd5b
|
DENN/MADD domain containing 5B
|
|
chr1_+_120285634
|
3.118
|
NM_001081276
NM_029709
NM_177548
|
Clasp1
|
CLIP associating protein 1
|
|
chr2_-_32286633
|
3.050
|
NM_001164358
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25
|
|
chr4_-_53172748
|
2.901
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr8_+_113443764
|
2.757
|
NM_009179
|
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr11_-_29147217
|
2.630
|
NM_025740
|
Ccdc104
|
coiled-coil domain containing 104
|
|
chr3_-_107499460
|
2.581
|
NM_145542
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1
|
|
chrX_+_38754480
|
2.578
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr2_+_35413977
|
2.531
|
NM_001114125
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr4_+_13711928
|
2.525
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_74416925
|
2.523
|
NM_001110209
NM_027133
|
Lnp
|
limb and neural patterns
|
|
chr15_-_81846328
|
2.456
|
NM_134095
|
Pppde2
|
PPPDE peptidase domain containing 2
|
|
chr9_-_101154160
|
2.436
|
NM_001161362
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr1_-_58642994
|
2.358
|
NM_172513
|
Fam126b
|
family with sequence similarity 126, member B
|
|
chr5_+_34916343
|
2.354
|
NM_001024458
NM_001102444
NM_013457
|
Add1
|
adducin 1 (alpha)
|
|
chr14_+_47621014
|
2.344
|
NM_001163433
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr2_-_37502738
|
2.293
|
NM_009261
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr10_-_126648509
|
2.259
|
NM_054097
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr3_-_115956312
|
2.244
|
NM_022427
|
Gpr88
|
G-protein coupled receptor 88
|
|
chr4_+_84530230
|
2.111
|
NM_175275
NM_177385
|
Cntln
|
centlein, centrosomal protein
|
|
chr3_-_116510948
|
2.089
|
NM_001081326
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase
|
|
chr4_+_97444312
|
2.083
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr4_+_13670425
|
2.033
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_21613162
|
1.968
|
NM_001039138
NM_001039139
NM_178597
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr19_-_37281762
|
1.905
|
NM_198300
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr6_-_38787222
|
1.883
|
NM_001136065
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr6_-_28084368
|
1.828
|
NM_008174
|
Grm8
|
glutamate receptor, metabotropic 8
|
|
chr5_+_43624701
|
1.725
|
NM_001177379
NM_175937
|
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr11_+_115243203
|
1.603
|
NM_001080929
|
Cdr2l
|
cerebellar degeneration-related protein 2-like
|
|
chr4_-_148001104
|
1.583
|
NM_001003898
NM_001003899
NM_001008545
NM_001008546
NM_145556
|
Tardbp
|
TAR DNA binding protein
|
|
chr2_-_7002344
|
1.557
|
NM_001110229
NM_001110230
NM_001160292
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr11_+_105451296
|
1.523
|
NM_181071
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr19_+_43514902
|
1.451
|
NM_031396
|
Cnnm1
|
cyclin M1
|
|
chr6_+_65621583
|
1.403
|
NM_172399
|
A930038C07Rik
|
RIKEN cDNA A930038C07 gene
|
|
chr5_+_58109246
|
1.389
|
NM_001122758
NM_018764
|
Pcdh7
|
protocadherin 7
|
|
chr15_+_88692581
|
1.324
|
NM_145478
|
Pim3
|
proviral integration site 3
|
|
chr1_+_174078145
|
1.309
|
NM_153555
|
Dcaf8
|
DDB1 and CUL4 associated factor 8
|
|
chr5_-_69732915
|
1.192
|
NM_175519
|
Kctd8
|
potassium channel tetramerisation domain containing 8
|
|
chr9_-_101101334
|
1.190
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr8_-_97400173
|
1.135
|
NM_053246
|
Dok4
|
docking protein 4
|
|
chr11_+_67508827
|
1.066
|
NM_009038
|
Rcvrn
|
recoverin
|
|
chr7_+_80520360
|
1.052
|
NM_177740
|
Rgma
|
RGM domain family, member A
|
|
chr11_-_84953442
|
1.051
|
NM_001029934
|
Usp32
|
ubiquitin specific peptidase 32
|
|
chr6_-_56312803
|
1.042
|
NM_001159960
|
Pde1c
|
phosphodiesterase 1C
|
|
chr14_+_21493519
|
1.020
|
NM_001168273
NM_172596
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr14_-_31166671
|
1.011
|
NM_001083616
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr12_-_109241423
|
0.914
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr3_+_135488454
|
0.817
|
NM_026228
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8
|
|
chr2_-_7317513
|
0.811
|
NM_001110232
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr9_-_42280256
|
0.782
|
NM_173038
|
Tbcel
|
tubulin folding cofactor E-like
|
|
chr19_+_38911056
|
0.760
|
NM_145952
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12
|
|
chr8_-_74009902
|
0.729
|
NM_001164679
|
Ano8
|
anoctamin 8
|
|
chr3_-_92674126
|
0.713
|
NM_025621
|
2310050C09Rik
|
RIKEN cDNA 2310050C09 gene
|
|
chr10_-_13194201
|
0.687
|
NM_001195065
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr9_+_65478364
|
0.584
|
NM_028030
|
Rbpms2
|
RNA binding protein with multiple splicing 2
|
|
chr11_-_45979836
|
0.563
|
NM_172524
|
Nipal4
|
NIPA-like domain containing 4
|
|
chr14_+_47502637
|
0.527
|
NM_001037221
NM_028966
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr6_-_56319574
|
0.526
|
NM_001159955
NM_001159956
NM_001159957
|
Pde1c
|
phosphodiesterase 1C
|
|
chr17_-_91492065
|
0.511
|
NM_020252
NM_177284
|
Nrxn1
|
neurexin I
|
|
chr5_-_90312358
|
0.509
|
NM_001081401
NM_177872
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3
|
|
chr5_+_111308801
|
0.469
|
NM_024477
|
Ttc28
|
tetratricopeptide repeat domain 28
|
|
chr10_+_98725858
|
0.465
|
NM_026268
|
Dusp6
|
dual specificity phosphatase 6
|
|
chr17_+_29169579
|
0.435
|
NM_013663
|
Srsf3
|
serine/arginine-rich splicing factor 3
|
|
chr6_-_56312170
|
0.402
|
NM_001025568
NM_001159952
NM_001159953
NM_011054
|
Pde1c
|
phosphodiesterase 1C
|
|
chr2_-_7317824
|
0.350
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr16_+_95923988
|
0.339
|
NM_011809
|
Ets2
|
E26 avian leukemia oncogene 2, 3' domain
|
|
chr17_-_24278482
|
0.305
|
NM_001080773
NM_011062
|
Pdpk1
|
3-phosphoinositide dependent protein kinase 1
|
|
chr5_+_13399301
|
0.295
|
NM_009152
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr1_-_52290058
|
0.234
|
NM_001081081
NM_001113383
|
Gls
|
glutaminase
|
|
chr1_-_80210403
|
0.228
|
NM_173425
|
Fam124b
|
family with sequence similarity 124, member B
|
|
chr9_-_78329444
|
0.222
|
NM_010106
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr7_+_54305973
|
0.152
|
NM_026436
|
Tmem86a
|
transmembrane protein 86A
|
|
chr6_-_38826156
|
0.120
|
NM_010433
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr1_+_181891219
|
0.098
|
NM_001033285
|
Cdc42bpa
|
CDC42 binding protein kinase alpha
|
|
chr2_+_90687308
|
0.049
|
NM_019758
|
Mtch2
|
mitochondrial carrier homolog 2 (C. elegans)
|
|
chr13_-_60278859
|
0.008
|
NM_008086
|
Gas1
|
growth arrest specific 1
|
|
chr11_+_35582969
|
0.003
|
NM_145962
|
Pank3
|
pantothenate kinase 3
|