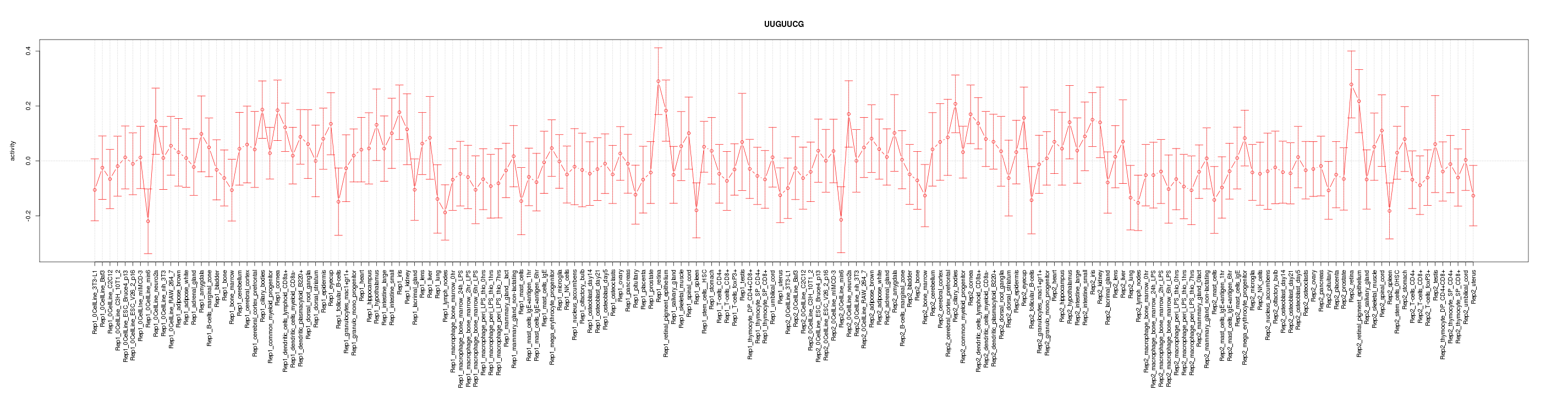
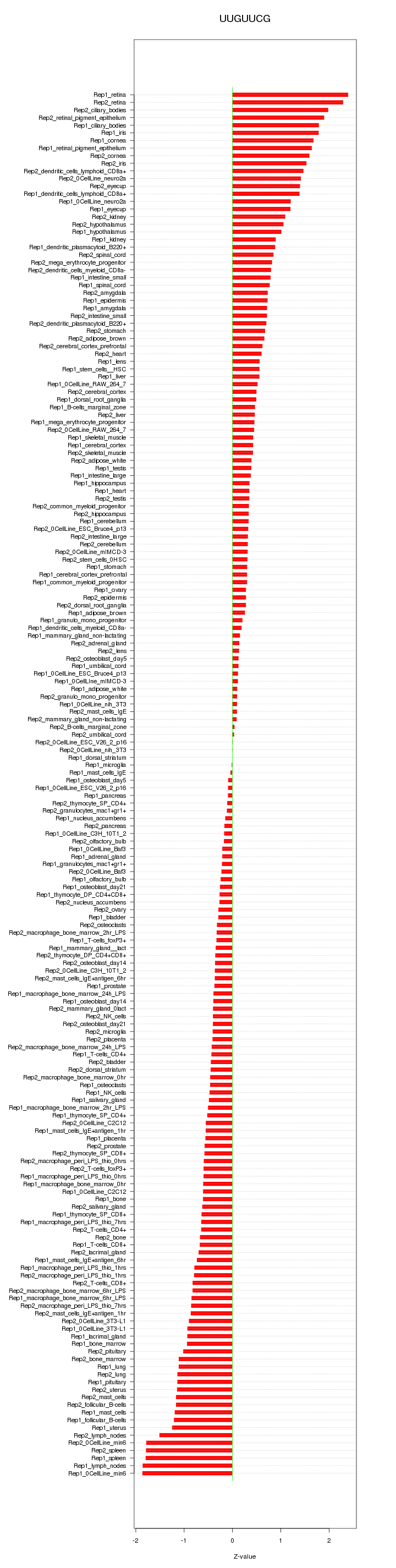
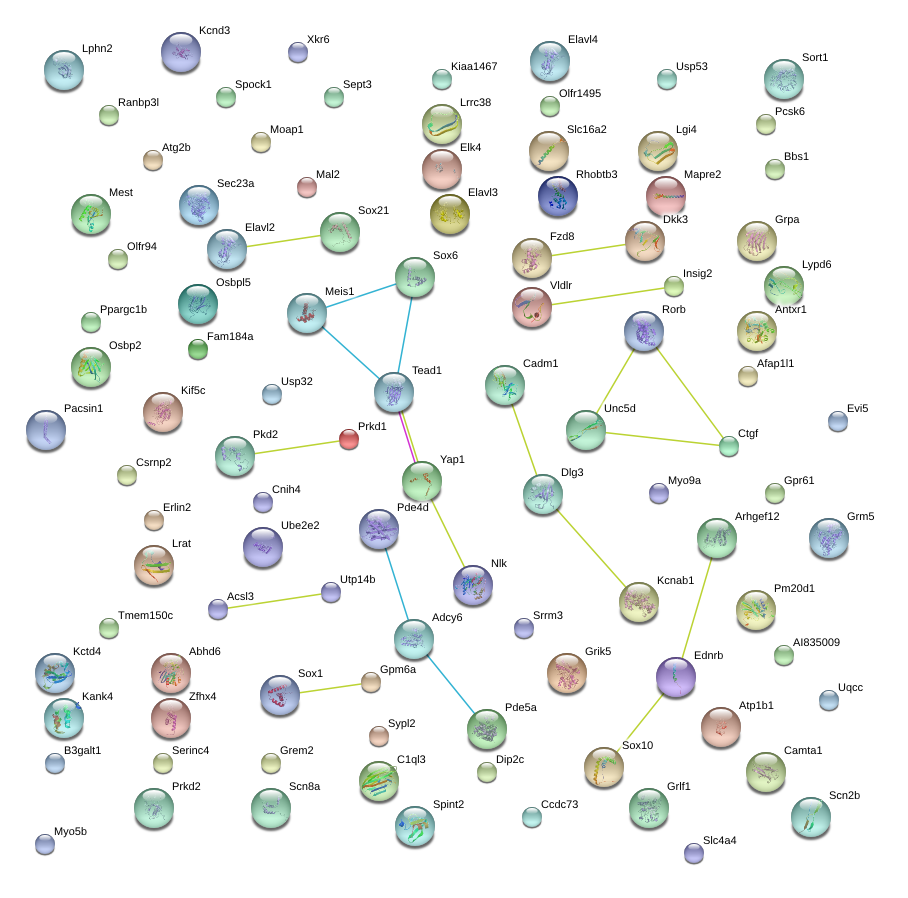
|
chr4_-_110024513
|
7.361
|
NM_001163397
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_-_119302546
|
7.003
|
NM_015814
|
Dkk3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr4_-_91038729
|
6.988
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_91066674
|
6.374
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_91042997
|
6.285
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_+_59598963
|
5.693
|
NM_173018
|
Myo9a
|
myosin IXa
|
|
chr4_-_109959366
|
5.652
|
NM_001163399
NM_010488
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr15_+_54402914
|
5.328
|
NM_178920
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr11_-_20731016
|
4.832
|
NM_173752
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr8_+_56040080
|
4.821
|
NM_153581
|
Gpm6a
|
glycoprotein m6a
|
|
chr3_+_5218545
|
4.625
|
NM_030708
|
Zfhx4
|
zinc finger homeodomain 4
|
|
chr18_-_61560053
|
4.503
|
NM_133249
|
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta
|
|
chr4_-_109960078
|
4.350
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_51750204
|
4.127
|
NM_008858
|
Prkd1
|
protein kinase D1
|
|
chr3_+_64913564
|
3.988
|
NM_010597
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr3_+_122432075
|
3.798
|
NM_153422
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific
|
|
chr1_-_166388431
|
3.784
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr19_-_4906607
|
3.749
|
NM_001033128
|
Bbs1
|
Bardet-Biedl syndrome 1 (human)
|
|
chr5_-_100588826
|
3.722
|
NM_182841
|
Tmem150c
|
transmembrane protein 150C
|
|
chr8_-_30330079
|
3.544
|
NM_153135
|
Unc5d
|
unc-5 homolog D (C. elegans)
|
|
chr15_+_82105322
|
3.536
|
NM_011889
|
Sept3
|
septin 3
|
|
chr19_+_27291507
|
3.527
|
NM_001161420
NM_013703
|
Vldlr
|
very low density lipoprotein receptor
|
|
chr14_-_19726121
|
3.519
|
NM_144839
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr1_-_123224303
|
3.512
|
NM_133748
|
Insig2
|
insulin induced gene 2
|
|
chr3_-_148617598
|
3.454
|
NM_001081298
|
Lphn2
|
latrophilin 2
|
|
chr1_+_78654399
|
3.445
|
NM_001001981
NM_001033606
NM_001136222
NM_028817
|
Utp14b
Acsl3
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog B (yeast)
acyl-CoA synthetase long-chain family member 3
|
|
chr19_-_19185685
|
3.413
|
NM_001043354
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr5_+_89316245
|
3.409
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr2_+_49921978
|
3.364
|
NM_177139
|
Lypd6
|
LY6/PLAUR domain containing 6
|
|
chr18_+_74602260
|
3.338
|
NM_201600
|
Myo5b
|
myosin VB
|
|
chr1_+_133693944
|
3.172
|
NM_178079
|
Pm20d1
|
peptidase M20 domain containing 1
|
|
chr14_+_8835385
|
3.166
|
NM_025341
|
Abhd6
|
abhydrolase domain containing 6
|
|
chrX_-_101017286
|
3.139
|
NM_009197
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2
|
|
chr13_-_58009653
|
3.107
|
NM_001166463
NM_001166464
NM_001166465
NM_001166466
NM_009262
|
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1
|
|
chr13_+_109444370
|
3.027
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr2_-_155755998
|
2.992
|
NM_018888
|
Uqcc
|
ubiquinol-cytochrome c reductase complex chaperone, CBP3 homolog (yeast)
|
|
chr7_-_123174560
|
2.956
|
NM_001025560
|
Sox6
|
SRY-box containing gene 6
|
|
chr14_+_64225366
|
2.935
|
NM_173393
|
Xkr6
|
X Kell blood group precursor related family member 6 homolog
|
|
chr6_-_87285690
|
2.912
|
NM_054041
|
Antxr1
|
anthrax toxin receptor 1
|
|
chr1_+_183081058
|
2.864
|
NM_030131
|
Cnih4
|
cornichon homolog 4 (Drosophila)
|
|
chr9_+_47338431
|
2.850
|
NM_001025600
NM_018770
NM_207675
NM_207676
|
Cadm1
|
cell adhesion molecule 1
|
|
chr15_+_100701068
|
2.781
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr14_-_118636251
|
2.735
|
NM_177753
|
Sox21
|
SRY-box containing gene 21
|
|
chr15_+_100766701
|
2.718
|
NM_011323
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr7_-_123138599
|
2.684
|
NM_011445
|
Sox6
|
SRY-box containing gene 6
|
|
chr17_+_27792587
|
2.677
|
NM_011861
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr12_-_60112987
|
2.625
|
NM_009147
|
Sec23a
|
SEC23A (S. cerevisiae)
|
|
chr3_-_108029499
|
2.595
|
NM_008596
|
Sypl2
|
synaptophysin-like 2
|
|
chrX_+_97972559
|
2.580
|
NM_001177780
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr6_+_135148004
|
2.574
|
NM_028982
|
8430419L09Rik
|
RIKEN cDNA 8430419L09 gene
|
|
chr18_+_9212853
|
2.573
|
NM_008058
|
Fzd8
|
frizzled homolog 8 (Drosophila)
|
|
chr2_+_67955769
|
2.558
|
NM_020283
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr7_-_17200285
|
2.531
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr7_+_94732677
|
2.451
|
NM_001081414
NM_001143834
|
Grm5
|
glutamate receptor, metabotropic 5
|
|
chr4_-_151235861
|
2.423
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr9_+_44925922
|
2.406
|
NM_001014761
|
Scn2b
|
sodium channel, voltage-gated, type II, beta
|
|
chr10_-_53470714
|
2.405
|
NM_001081428
|
Fam184a
|
family with sequence similarity 184, member A
|
|
chr8_+_128193717
|
2.381
|
NM_028908
|
4933403G14Rik
|
RIKEN cDNA 4933403G14 gene
|
|
chr5_+_104888468
|
2.368
|
NM_008861
|
Pkd2
|
polycystic kidney disease 2
|
|
chr11_-_18918923
|
2.345
|
NM_001193271
NM_010789
|
Meis1
|
Meis homeobox 1
|
|
chr14_-_104243379
|
2.344
|
NM_001136061
|
Ednrb
|
endothelin receptor type B
|
|
chr14_+_76354807
|
2.338
|
NM_026214
|
Kctd4
|
potassium channel tetramerisation domain containing 4
|
|
chr6_+_30688050
|
2.295
|
NM_008590
|
Mest
|
mesoderm specific transcript
|
|
chr13_+_9275741
|
2.291
|
NM_001081426
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr12_-_103981869
|
2.252
|
NM_001142937
NM_022323
|
Moap1
|
modulator of apoptosis 1
|
|
chr18_+_23962468
|
2.195
|
NM_153058
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_-_3763899
|
2.125
|
NM_152818
|
Osbp2
|
oxysterol binding protein 2
|
|
chr2_-_12932442
|
2.125
|
NM_153155
|
C1ql3
|
C1q-like 3
|
|
chr11_-_78510874
|
2.109
|
NM_008702
|
Nlk
|
nemo like kinase
|
|
chr2_+_49474775
|
2.060
|
NM_008449
|
Kif5c
|
kinesin family member 5C
|
|
chr12_-_106923394
|
2.046
|
NM_029654
|
Atg2b
|
ATG2 autophagy related 2 homolog B (S. cerevisiae)
|
|
chr5_+_136293975
|
2.032
|
NM_021403
|
Srrm3
|
serine/arginine repetitive matrix 3
|
|
chr9_-_21856445
|
2.000
|
NM_010487
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr1_-_176851949
|
1.975
|
NM_011825
|
Grem2
|
gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis)
|
|
chr11_-_84953442
|
1.957
|
NM_001029934
|
Usp32
|
ubiquitin specific peptidase 32
|
|
chr1_+_133904162
|
1.952
|
NM_007923
|
Elk4
|
ELK4, member of ETS oncogene family
|
|
chr4_+_142939652
|
1.942
|
NM_001162983
|
Lrrc38
|
leucine rich repeat containing 38
|
|
chr2_-_121282428
|
1.926
|
NM_001025371
|
Serinc4
|
serine incorporator 4
|
|
chr3_-_122687238
|
1.914
|
NM_133857
|
Usp53
|
ubiquitin specific peptidase 53
|
|
chrX_+_97969784
|
1.894
|
NM_001177779
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr5_-_108304062
|
1.891
|
NM_007964
|
Evi5
|
ecotropic viral integration site 5
|
|
chr15_-_98438063
|
1.878
|
NM_007405
|
Adcy6
|
adenylate cyclase 6
|
|
chr3_+_105255247
|
1.863
|
NM_001039347
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3
|
|
chr3_-_107957795
|
1.862
|
NM_175470
|
Gpr61
|
G protein-coupled receptor 61
|
|
chr9_-_42913797
|
1.840
|
NM_027144
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr7_-_25857275
|
1.833
|
NM_008168
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2)
|
|
chrX_+_97963061
|
1.832
|
NM_001177778
NM_016747
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr15_-_100325669
|
1.805
|
NM_153407
|
Csrnp2
|
cysteine-serine-rich nuclear protein 2
|
|
chr13_-_76081244
|
1.805
|
NM_028493
|
Rhobtb3
|
Rho-related BTB domain containing 3
|
|
chr8_+_28134259
|
1.802
|
NM_153592
|
Erlin2
|
ER lipid raft associated 2
|
|
chr3_-_82707880
|
1.788
|
NM_023624
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase)
|
|
chr15_+_8918001
|
1.787
|
NM_198024
|
Ranbp3l
|
RAN binding protein 3-like
|
|
chr4_-_98484163
|
1.754
|
NM_172872
|
Kank4
|
KN motif and ankyrin repeat domains 4
|
|
chr7_+_31844947
|
1.748
|
NM_144556
|
Lgi4
|
leucine-rich repeat LGI family, member 4
|
|
chr7_+_73007017
|
1.737
|
NM_011048
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr18_-_61946299
|
1.732
|
NM_178928
|
Afap1l1
|
actin filament associated protein 1-like 1
|
|
chr10_+_24315247
|
1.732
|
NM_010217
|
Ctgf
|
connective tissue growth factor
|
|
chr9_-_8004559
|
1.716
|
NM_001171147
NM_009534
|
Yap1
|
yes-associated protein 1
|
|
chr19_-_19075535
|
1.687
|
NM_146095
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr7_+_119822831
|
1.681
|
NM_001166584
NM_009346
|
Tead1
|
TEA domain family member 1
|
|
chr2_+_104726480
|
1.679
|
NM_177600
|
Ccdc73
|
coiled-coil domain containing 73
|
|
chr3_+_108087046
|
1.677
|
NM_019972
|
Sort1
|
sortilin 1
|
|
chr7_+_119902897
|
1.671
|
NM_001166585
|
Tead1
|
TEA domain family member 1
|
|
chr16_-_94509462
|
1.661
|
NM_139145
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase)
|
|
chr13_-_52626065
|
1.641
|
NM_001024474
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr4_+_43071852
|
1.631
|
NM_001081413
NM_021468
|
Unc13b
|
unc-13 homolog B (C. elegans)
|
|
chr7_-_123182257
|
1.598
|
NM_001025559
|
Sox6
|
SRY-box containing gene 6
|
|
chr10_+_19654195
|
1.581
|
NM_008580
|
Map3k5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chrX_+_130593867
|
1.577
|
NM_133196
|
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2
|
|
chr12_+_21117592
|
1.573
|
NM_001004364
NM_001098168
NM_001135192
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr14_-_104242892
|
1.572
|
NM_007904
|
Ednrb
|
endothelin receptor type B
|
|
chr9_+_122714664
|
1.570
|
NM_001013405
|
D9Ertd402e
|
DNA segment, Chr 9, ERATO Doi 402, expressed
|
|
chr16_+_29209780
|
1.566
|
NM_013751
|
Hrasls
|
HRAS-like suppressor
|
|
chr15_+_97998090
|
1.562
|
NM_177716
|
AI836003
|
expressed sequence AI836003
|
|
chr15_-_77949709
|
1.549
|
NM_007583
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr1_+_9591347
|
1.536
|
NM_178399
|
3110035E14Rik
|
RIKEN cDNA 3110035E14 gene
|
|
chr2_+_34287544
|
1.502
|
NM_177345
|
Mapkap1
|
mitogen-activated protein kinase associated protein 1
|
|
chr6_-_14705104
|
1.462
|
NM_080464
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr2_-_103143352
|
1.461
|
NM_007914
|
Ehf
|
ets homologous factor
|
|
chrX_+_137212566
|
1.460
|
NM_011845
|
Mid2
|
midline 2
|
|
chr6_-_30643681
|
1.451
|
NM_031998
|
Tsga14
|
testis specific gene A14
|
|
chr2_-_136213509
|
1.450
|
NM_172858
|
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr15_-_58654964
|
1.433
|
NM_175212
|
Tmem65
|
transmembrane protein 65
|
|
chr15_-_100382354
|
1.428
|
NM_033476
|
Tcfcp2
|
transcription factor CP2
|
|
chr7_-_140315100
|
1.427
|
NM_009980
|
Ctbp2
|
C-terminal binding protein 2
|
|
chr6_+_67216948
|
1.426
|
NM_001113564
NM_001113565
NM_001113566
NM_025814
|
Serbp1
|
serpine1 mRNA binding protein 1
|
|
chr13_-_64349830
|
1.373
|
NM_001122989
|
Cdc14b
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chrX_+_157205944
|
1.361
|
NM_011302
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr14_-_63864060
|
1.357
|
NM_008092
|
Gata4
|
GATA binding protein 4
|
|
chr7_+_94395160
|
1.347
|
NM_015760
|
Nox4
|
NADPH oxidase 4
|
|
chr1_-_64784316
|
1.345
|
NM_001042659
NM_022721
|
Fzd5
|
frizzled homolog 5 (Drosophila)
|
|
chr12_+_79327630
|
1.334
|
NM_145965
NM_172952
|
Gphn
|
gephyrin
|
|
chr15_+_102864825
|
1.321
|
NM_013553
|
Hoxc4
|
homeobox C4
|
|
chr14_+_67852128
|
1.314
|
NM_010095
|
Ebf2
|
early B-cell factor 2
|
|
chr15_-_98493302
|
1.314
|
NM_001080981
|
Ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr16_-_37385024
|
1.304
|
NM_001114611
NM_001114612
NM_001114613
NM_172440
|
Stxbp5l
|
syntaxin binding protein 5-like
|
|
chr2_+_169459145
|
1.265
|
NM_080455
|
Tshz2
|
teashirt zinc finger family member 2
|
|
chr14_+_67530044
|
1.251
|
NM_175498
|
Pnma2
|
paraneoplastic antigen MA2
|
|
chr11_-_63735652
|
1.251
|
NM_018805
|
Hs3st3b1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr6_-_52163065
|
1.250
|
NM_010452
|
Hoxa3
|
homeobox A3
|
|
chr1_+_106665395
|
1.244
|
NM_011800
|
Cdh20
|
cadherin 20
|
|
chr8_-_128016609
|
1.202
|
NM_001081337
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2
|
|
chr7_+_148317160
|
1.201
|
NM_013782
|
Ptdss2
|
phosphatidylserine synthase 2
|
|
chr10_-_85961630
|
1.200
|
NM_001164495
NM_013722
|
Syn3
|
synapsin III
|
|
chr16_+_31428833
|
1.197
|
NM_001122683
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr6_-_28084368
|
1.179
|
NM_008174
|
Grm8
|
glutamate receptor, metabotropic 8
|
|
chr3_+_94114044
|
1.167
|
NM_029431
|
Them4
|
thioesterase superfamily member 4
|
|
chr1_-_182126032
|
1.153
|
NM_023341
|
Adck3
|
aarF domain containing kinase 3
|
|
chr13_-_40825789
|
1.151
|
NM_001122948
|
Tcfap2a
|
transcription factor AP-2, alpha
|
|
chr15_-_58765248
|
1.148
|
NM_175151
|
Tatdn1
|
TatD DNase domain containing 1
|
|
chr3_-_141645214
|
1.134
|
NM_007560
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B
|
|
chr2_+_105516601
|
1.129
|
NM_013627
|
Pax6
|
paired box gene 6
|
|
chr18_+_42435006
|
1.126
|
NM_172626
|
Rbm27
|
RNA binding motif protein 27
|
|
chr2_-_52532100
|
1.110
|
NM_001037099
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr8_+_125930981
|
1.109
|
NM_008559
|
Mc1r
|
melanocortin 1 receptor
|
|
chr11_-_88725877
|
1.103
|
NM_001042541
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr7_+_107855215
|
1.089
|
NM_178764
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr13_+_25147872
|
1.088
|
NM_001195617
NM_177577
|
Dcdc2a
|
doublecortin domain containing 2a
|
|
chr16_+_31422377
|
1.085
|
NM_175177
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr15_+_40486577
|
1.079
|
NM_011766
|
Zfpm2
|
zinc finger protein, multitype 2
|
|
chr2_+_163820920
|
1.078
|
NM_018753
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chrX_+_6356431
|
1.074
|
NM_177914
|
Dgkk
|
diacylglycerol kinase kappa
|
|
chr13_-_64376186
|
1.059
|
NM_172587
|
Cdc14b
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_+_153602492
|
1.053
|
NM_001039644
|
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr12_-_103995925
|
1.052
|
NM_001142938
NM_001142939
|
AK010878
|
cDNA sequence AK010878
|
|
chrX_+_138662479
|
1.041
|
NM_001161430
|
Nxt2
|
nuclear transport factor 2-like export factor 2
|
|
chr2_-_52414072
|
1.009
|
NM_146123
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr11_-_88712812
|
0.993
|
NM_009648
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr14_+_47621014
|
0.988
|
NM_001163433
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr10_+_110735734
|
0.988
|
NM_027914
|
Bbs10
|
Bardet-Biedl syndrome 10 (human)
|
|
chr1_-_182123596
|
0.987
|
NM_001163290
|
Adck3
|
aarF domain containing kinase 3
|
|
chr10_+_128185549
|
0.975
|
NM_001170869
|
Wibg
|
within bgcn homolog (Drosophila)
|
|
chr11_+_115281434
|
0.972
|
NM_183285
|
Kctd2
|
potassium channel tetramerisation domain containing 2
|
|
chr7_-_144506112
|
0.968
|
NM_001113414
NM_001113415
NM_010096
|
Ebf3
|
early B-cell factor 3
|
|
chr7_+_13466525
|
0.956
|
NM_153802
|
Zfp128
|
zinc finger protein 128
|
|
chr5_+_43624701
|
0.951
|
NM_001177379
NM_175937
|
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr7_-_135020290
|
0.929
|
NM_146259
|
Zfp668
|
zinc finger protein 668
|
|
chr2_-_77654669
|
0.925
|
NM_178723
|
Zfp385b
|
zinc finger protein 385B
|
|
chr10_-_49508472
|
0.911
|
NM_001111268
NM_010349
|
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2)
|
|
chr5_+_67651890
|
0.911
|
NM_028975
NM_030108
|
Tmem33
|
transmembrane protein 33
|
|
chrX_+_138662725
|
0.878
|
NM_172782
|
Nxt2
|
nuclear transport factor 2-like export factor 2
|
|
chr8_-_114400574
|
0.866
|
NM_025781
|
Tmem170
|
transmembrane protein 170
|
|
chr9_-_79641213
|
0.859
|
NM_133718
|
Tmem30a
|
transmembrane protein 30A
|
|
chr15_+_89330287
|
0.858
|
NM_021423
|
Shank3
|
SH3/ankyrin domain gene 3
|
|
chr13_-_48720941
|
0.836
|
NM_207232
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1
|
|
chr3_+_80840337
|
0.820
|
NM_019971
|
Pdgfc
|
platelet-derived growth factor, C polypeptide
|
|
chr2_-_77541328
|
0.802
|
NM_001113399
|
Zfp385b
|
zinc finger protein 385B
|
|
chr15_-_90509751
|
0.798
|
NM_001033851
NM_025815
|
Cpne8
|
copine VIII
|
|
chr8_+_84239098
|
0.798
|
NM_001024617
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II
|
|
chr5_-_24606422
|
0.791
|
NM_145401
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr17_-_82137708
|
0.782
|
NM_001112798
NM_011406
|
Slc8a1
D830013E24Rik
|
solute carrier family 8 (sodium/calcium exchanger), member 1
RIKEN cDNA D830013E24 gene
|
|
chrX_+_98644445
|
0.776
|
NM_013712
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr6_-_118147735
|
0.757
|
NM_001080780
NM_009050
|
Ret
|
ret proto-oncogene
|
|
chr18_+_69505374
|
0.751
|
NM_001083967
|
Tcf4
|
transcription factor 4
|
|
chr2_+_62502329
|
0.747
|
NM_145523
|
Gca
|
grancalcin
|
|
chr2_-_38781939
|
0.743
|
NM_010264
|
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr2_-_77357674
|
0.724
|
NM_001113400
|
Zfp385b
|
zinc finger protein 385B
|
|
chr7_+_96552871
|
0.722
|
NM_008055
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr1_-_138850206
|
0.717
|
NM_001159769
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr10_-_112365976
|
0.716
|
NM_001033474
|
Atxn7l3b
|
ataxin 7-like 3B
|