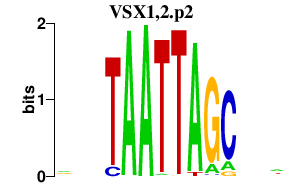
|
chr10_+_97028409
|
11.883
|
|
Lum
|
lumican
|
|
chr2_+_20211539
|
6.004
|
NM_001081006
|
Etl4
|
enhancer trap locus 4
|
|
chr4_+_133624366
|
4.680
|
NM_001162970
|
Aim1l
|
absent in melanoma 1-like
|
|
chr3_-_27316628
|
3.767
|
|
Fndc3b
|
fibronectin type III domain containing 3B
|
|
chr9_-_104028940
|
3.286
|
NM_145700
|
Acad11
Ccrl1
|
acyl-Coenzyme A dehydrogenase family, member 11
chemokine (C-C motif) receptor-like 1
|
|
chr4_-_14548903
|
2.922
|
NM_145947
|
Slc26a7
|
solute carrier family 26, member 7
|
|
chr1_-_72920776
|
2.396
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr2_-_168592528
|
2.340
|
NM_175303
NM_201395
NM_201396
|
Sall4
|
sal-like 4 (Drosophila)
|
|
chr19_+_38207270
|
2.259
|
NM_001170959
NM_033614
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime
|
|
chr10_-_26095736
|
2.251
|
|
L3mbtl3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr7_+_31536489
|
2.169
|
NM_001083903
NM_172205
|
Sbsn
|
suprabasin
|
|
chr8_+_11448095
|
2.168
|
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr8_-_11199802
|
2.097
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr8_+_46744023
|
1.860
|
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_140019846
|
1.752
|
NM_013702
|
Uncx
|
UNC homeobox
|
|
chr4_+_145260599
|
1.588
|
NM_001103158
|
Gm13242
|
predicted gene 13242
|
|
chr2_-_25325248
|
1.533
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr11_+_60351159
|
1.530
|
NM_172943
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr6_+_77193078
|
1.512
|
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chrX_+_16196823
|
1.494
|
NM_173740
|
Maoa
|
monoamine oxidase A
|
|
chr5_-_113510602
|
1.237
|
NM_001159650
NM_021352
|
Crybb3
|
crystallin, beta B3
|
|
chr9_+_43118845
|
1.091
|
NM_023655
|
Trim29
|
tripartite motif-containing 29
|
|
chr4_+_145850602
|
1.007
|
NM_001177767
NM_009051
|
Gm13138
Rex2
|
predicted gene 13138
reduced expression 2
|
|
chr4_+_146395958
|
1.007
|
NM_001177767
NM_009051
|
Gm13138
Rex2
|
predicted gene 13138
reduced expression 2
|
|
chr7_-_141568510
|
0.974
|
NM_001081331
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed
|
|
chr4_+_146124224
|
0.961
|
NM_001177546
|
Zfp600
|
zinc finger protein 600
|
|
chr10_+_39332934
|
0.921
|
|
Traf3ip2
|
TRAF3 interacting protein 2
|
|
chr5_-_86802390
|
0.920
|
NM_145561
|
Tmprss11d
|
transmembrane protease, serine 11d
|
|
chr6_-_136730254
|
0.889
|
NM_001127318
NM_145067
|
Gucy2c
|
guanylate cyclase 2c
|
|
chr16_+_45094196
|
0.847
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr18_+_57628139
|
0.839
|
NM_001134697
|
Ctxn3
|
cortexin 3
|
|
chr3_+_159502646
|
0.826
|
NM_026582
|
Wls
|
wntless homolog (Drosophila)
|
|
chr3_-_49561168
|
0.807
|
NM_130448
|
Pcdh18
|
protocadherin 18
|
|
chr9_+_31923720
|
0.796
|
NM_001195632
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr7_-_123174560
|
0.778
|
NM_001025560
|
Sox6
|
SRY-box containing gene 6
|
|
chr9_-_91255831
|
0.777
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr2_+_121184334
|
0.753
|
NM_009897
|
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous
|
|
chr1_+_24184448
|
0.742
|
NM_007740
|
Col9a1
|
collagen, type IX, alpha 1
|
|
chr10_+_97110043
|
0.727
|
|
Epyc
|
epiphycan
|
|
chr6_-_68967061
|
0.723
|
|
|
|
|
chr18_-_35881144
|
0.691
|
NM_001033141
|
Ecscr
|
endothelial cell-specific chemotaxis regulator
|
|
chr7_+_53631843
|
0.682
|
NM_010866
|
Myod1
|
myogenic differentiation 1
|
|
chr10_+_39332739
|
0.676
|
NM_134000
|
Traf3ip2
|
TRAF3 interacting protein 2
|
|
chr6_-_69367406
|
0.673
|
|
Igkv4-68
|
immunoglobulin kappa variable 4-68
|
|
chr10_+_73284612
|
0.672
|
NM_001142735
NM_001142736
NM_001142737
NM_001142738
NM_001142739
NM_001142740
NM_001142741
NM_001142742
NM_001142743
NM_001142746
NM_001142747
NM_001142748
NM_001142760
NM_023115
|
Pcdh15
|
protocadherin 15
|
|
chr6_-_69494873
|
0.656
|
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1
|
|
chr5_+_90919768
|
0.634
|
|
Afp
|
alpha fetoprotein
|
|
chr5_+_90919758
|
0.634
|
|
Afp
|
alpha fetoprotein
|
|
chr12_-_105711324
|
0.626
|
NM_010351
|
Gsc
|
goosecoid homeobox
|
|
chr4_-_147016606
|
0.616
|
NM_145078
|
2610305D13Rik
|
RIKEN cDNA 2610305D13 gene
|
|
chr2_+_103909961
|
0.567
|
|
Cd59b
|
CD59b antigen
|
|
chr1_-_72921098
|
0.552
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr1_+_174486066
|
0.547
|
NM_177723
|
Vsig8
|
V-set and immunoglobulin domain containing 8
|
|
chr1_+_40381420
|
0.537
|
NM_133193
|
Il1rl2
|
interleukin 1 receptor-like 2
|
|
chr6_-_130287518
|
0.537
|
NM_010648
|
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3
|
|
chr17_+_17539832
|
0.533
|
|
Lix1
|
limb expression 1 homolog (chicken)
|
|
chr12_-_11215753
|
0.528
|
NM_019544
|
Msgn1
|
mesogenin 1
|
|
chr19_+_56798916
|
0.478
|
|
Adrb1
|
adrenergic receptor, beta 1
|
|
chr5_-_84846348
|
0.465
|
NM_007937
|
Epha5
|
Eph receptor A5
|
|
chr12_+_110783786
|
0.462
|
|
Meg3
|
maternally expressed 3
|
|
chr2_+_151736942
|
0.403
|
NM_009641
|
Angpt4
|
angiopoietin 4
|
|
chr6_-_116666759
|
0.389
|
NM_178768
|
Tmem72
|
transmembrane protein 72
|
|
chr2_+_3341402
|
0.344
|
NM_001110214
NM_146114
NM_175683
|
Dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae)
|
|
chr13_-_33250219
|
0.341
|
NM_011453
|
Serpinb9c
|
serine (or cysteine) peptidase inhibitor, clade B, member 9c
|
|
chr5_+_90919739
|
0.334
|
NM_007423
|
Afp
|
alpha fetoprotein
|
|
chr6_+_77192839
|
0.329
|
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr4_-_147183894
|
0.314
|
NM_001127189
|
Gm13157
2610036A22Rik
|
predicted gene 13157
RIKEN cDNA 2610036A22 gene
|
|
chr11_-_65083322
|
0.307
|
|
Myocd
|
myocardin
|
|
chr15_-_8660729
|
0.281
|
NM_148938
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr6_+_57063418
|
0.281
|
NM_053231
|
Vmn1r10
|
vomeronasal 1 receptor 10
|
|
chr11_-_99184254
|
0.278
|
NM_133730
|
Krt25
|
keratin 25
|
|
chr4_+_146927385
|
0.268
|
NM_001014397
|
Gm13154
|
predicted gene 13154
|
|
chr5_-_91069499
|
0.259
|
NM_028478
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr6_-_130143018
|
0.246
|
NM_010651
|
Klra5
Klra6
Klra9
|
killer cell lectin-like receptor, subfamily A, member 5
killer cell lectin-like receptor, subfamily A, member 6
killer cell lectin-like receptor subfamily A, member 9
|
|
chr1_-_132586370
|
0.238
|
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr2_-_164410946
|
0.234
|
NM_001033240
|
Wfdc6a
|
WAP four-disulfide core domain 6A
|
|
chr11_-_48864726
|
0.231
|
NM_173434
|
9930111J21Rik2
|
RIKEN cDNA 9930111J21 gene 2
|
|
chr3_-_130433225
|
0.230
|
NM_026724
|
Rpl34-ps1
Rpl34
|
ribosomal protein L34, pseudogene 1
ribosomal protein L34
|
|
chr4_-_132066370
|
0.217
|
NM_144907
|
Sesn2
|
sestrin 2
|
|
chr10_+_98453513
|
0.211
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr12_+_56699903
|
0.201
|
NM_020287
|
Insm2
|
insulinoma-associated 2
|
|
chr17_+_17539908
|
0.191
|
|
Lix1
|
limb expression 1 homolog (chicken)
|
|
chr11_-_94693315
|
0.186
|
NM_001163172
|
Tmem92-ps
|
transmembrane protein 92, pseudogene
|
|
chr17_+_35670145
|
0.183
|
NM_020576
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human)
|
|
chr5_-_84846256
|
0.175
|
|
Epha5
|
Eph receptor A5
|
|
chr10_+_98725871
|
0.171
|
|
Dusp6
|
dual specificity phosphatase 6
|
|
chr15_+_101303908
|
0.160
|
NM_010667
|
Krt86
|
keratin 86
|
|
chr3_+_92748407
|
0.154
|
NM_033175
|
Lce3c
|
late cornified envelope 3C
|
|
chr16_-_92400226
|
0.138
|
NM_019466
|
Rcan1
|
regulator of calcineurin 1
|
|
chr10_+_127335573
|
0.118
|
NM_027301
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7
|
|
chr15_-_101573522
|
0.118
|
NM_019956
|
Krt71
|
keratin 71
|
|
chr4_+_129138924
|
0.110
|
|
Bsdc1
|
BSD domain containing 1
|
|
chr6_+_57087346
|
0.107
|
NM_053233
|
Vmn1r11
|
vomeronasal 1 receptor 11
|
|
chr19_-_55315725
|
0.104
|
NM_001081076
|
Gucy2g
|
guanylate cyclase 2g
|
|
chr15_-_101632762
|
0.096
|
NM_212485
|
Krt73
|
keratin 73
|
|
chr15_+_4677207
|
0.093
|
NM_016704
|
C6
|
complement component 6
|
|
chr11_-_69648350
|
0.088
|
NM_198862
|
Nlgn2
|
neuroligin 2
|
|
chr13_-_16115192
|
0.086
|
|
B230303A05Rik
|
RIKEN cDNA B230303A05 gene
|
|
chr10_-_106922986
|
0.064
|
NM_008656
|
Myf5
|
myogenic factor 5
|
|
chr17_+_26792031
|
0.058
|
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr3_+_93272675
|
0.053
|
NM_027762
|
Tchhl1
|
trichohyalin-like 1
|
|
chr6_+_57020888
|
0.032
|
NM_134185
|
Vmn1r9
|
vomeronasal 1 receptor 9
|
|
chr3_-_115956312
|
0.019
|
NM_022427
|
Gpr88
|
G-protein coupled receptor 88
|
|
chrM_+_13543
|
0.012
|
|
|
|
|
chr4_+_110070336
|
0.008
|
NM_001048189
NM_030231
|
Agbl4
|
ATP/GTP binding protein-like 4
|
|
chr1_+_179659944
|
0.005
|
NM_027077
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene
|