|
chr9_-_117161234
|
45.531
|
|
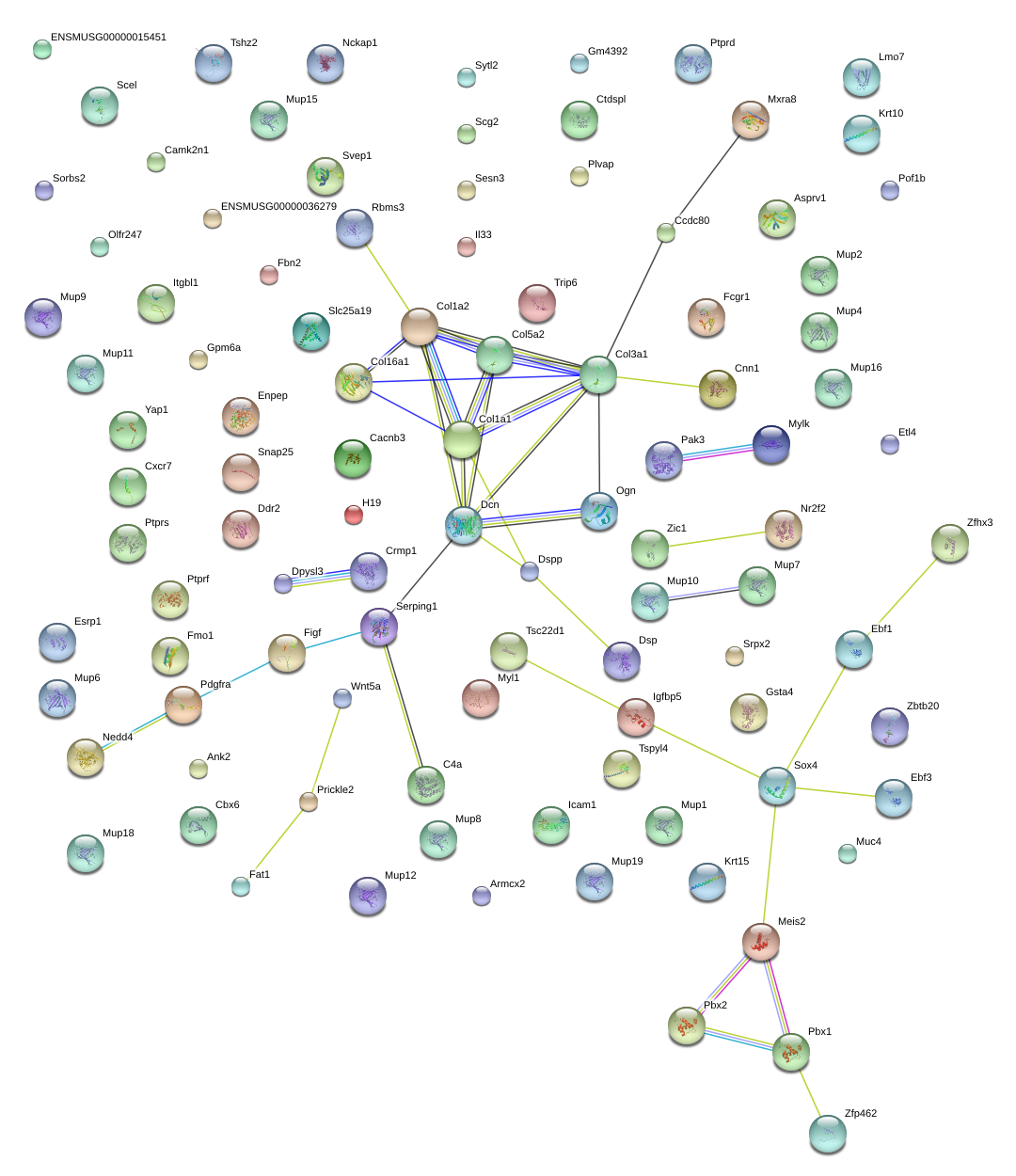
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr9_-_117161564
|
35.136
|
NM_001172122
NM_001172123
NM_001172124
NM_001172126
NM_178660
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr9_-_117160916
|
32.414
|
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr1_-_45559866
|
24.121
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr11_-_55208276
|
24.088
|
|
|
|
|
chr6_+_4484651
|
22.152
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr13_+_49704471
|
20.585
|
|
Ogn
|
osteoglycin
|
|
chr7_+_97529155
|
19.171
|
NM_001040086
|
Sytl2
|
synaptotagmin-like 2
|
|
chr1_-_45559904
|
19.047
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_+_45368443
|
18.383
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr2_-_115890747
|
17.880
|
NM_001136072
NM_001159567
NM_001159568
NM_010825
|
Meis2
|
Meis homeobox 2
|
|
chr16_+_34921598
|
17.225
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr1_-_170362298
|
17.049
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr18_-_43552694
|
16.801
|
NM_009468
|
Dpysl3
|
dihydropyrimidinase-like 3
|
|
chr18_-_58169723
|
16.609
|
|
Fbn2
|
fibrillin 2
|
|
chr1_-_45560001
|
16.486
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_-_45559700
|
16.060
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr6_+_4490515
|
15.782
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr4_+_55093796
|
15.285
|
|
Zfp462
|
zinc finger protein 462
|
|
chrX_+_160811667
|
15.156
|
|
Figf
|
c-fos induced growth factor
|
|
chr16_+_34995000
|
14.989
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr5_+_75548309
|
14.859
|
NM_011058
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr1_-_170085342
|
14.466
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr1_-_170085798
|
14.390
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr6_+_4465627
|
12.518
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr4_-_11313886
|
12.437
|
NM_194055
|
Esrp1
|
epithelial splicing regulatory protein 1
|
|
chr1_+_45368382
|
12.421
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_-_170362388
|
12.287
|
NM_008783
NM_183355
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr1_-_164796609
|
12.283
|
|
Fmo1
|
flavin containing monooxygenase 1
|
|
chr1_-_164796660
|
12.220
|
NM_010231
|
Fmo1
|
flavin containing monooxygenase 1
|
|
chr10_+_96942181
|
12.184
|
|
Dcn
|
decorin
|
|
chr16_+_43619057
|
12.046
|
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr9_+_72584532
|
11.932
|
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr11_-_99997241
|
11.765
|
NM_008469
|
Krt15
|
keratin 15
|
|
chr4_-_61179982
|
11.759
|
|
Mup1
|
major urinary protein 1
|
|
chr2_-_84615535
|
11.726
|
NM_009776
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr8_+_46744023
|
11.680
|
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr8_+_46035561
|
11.651
|
NM_001081286
|
Fat1
|
FAT tumor suppressor homolog 1 (Drosophila)
|
|
chr9_+_78039765
|
11.463
|
NM_010357
|
Gsta4
|
glutathione S-transferase, alpha 4
|
|
chr5_+_75548337
|
11.347
|
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr19_+_29999596
|
11.124
|
NM_133775
|
Il33
|
interleukin 33
|
|
chr9_+_92502076
|
11.103
|
|
|
|
|
chr1_-_72905107
|
10.967
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr1_+_92100572
|
10.826
|
NM_007722
|
Cxcr7
|
chemokine (C-X-C motif) receptor 7
|
|
chr1_-_79436564
|
10.318
|
NM_009129
|
Scg2
|
secretogranin II
|
|
chr15_-_79656745
|
10.302
|
|
Cbx6
|
chromobox homolog 6
|
|
chr9_+_21903702
|
9.965
|
|
Cnn1
|
calponin 1
|
|
chr6_-_92431385
|
9.962
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chrX_-_109812258
|
9.865
|
NM_181579
|
Pof1b
|
premature ovarian failure 1B
|
|
chr1_-_170361525
|
9.734
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr8_+_111482660
|
9.667
|
|
Zfhx3
|
zinc finger homeobox 3
|
|
chrX_+_130442964
|
9.612
|
NM_001083895
NM_026838
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2
|
|
chr9_+_21903696
|
9.603
|
NM_009922
|
Cnn1
|
calponin 1
|
|
chr7_-_77505248
|
9.438
|
NM_009697
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_+_169459145
|
9.435
|
NM_080455
|
Tshz2
|
teashirt zinc finger family member 2
|
|
chr9_-_91260619
|
9.263
|
NM_009573
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chrX_+_160811458
|
9.008
|
NM_010216
|
Figf
|
c-fos induced growth factor
|
|
chr16_+_32735971
|
8.998
|
NM_080457
|
Muc4
|
mucin 4
|
|
chr13_-_29043617
|
8.884
|
|
Sox4
|
SRY-box containing gene 4
|
|
chr4_-_61335031
|
8.750
|
|
Mup10
LOC100048884
Mup19
Mup1
Mup2
|
major urinary protein 10
novel member of the major urinary protein (Mup) gene family
major urinary protein 19
major urinary protein 1
major urinary protein 2
|
|
chr10_+_96942124
|
8.748
|
NM_001190451
|
Dcn
|
decorin
|
|
chr11_+_44431601
|
8.568
|
NM_007897
|
Ebf1
|
early B-cell factor 1
|
|
chr4_-_58083082
|
8.539
|
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr10_+_34018323
|
8.465
|
|
Tspyl4
|
TSPY-like 4
|
|
chr14_+_29318658
|
8.386
|
NM_009524
|
Wnt5a
|
wingless-related MMTV integration site 5A
|
|
chr4_+_155213788
|
8.362
|
NM_024263
|
Mxra8
|
matrix-remodelling associated 8
|
|
chr16_+_45094196
|
8.345
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr1_-_170361985
|
8.179
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr6_+_86578225
|
8.173
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr11_+_94812919
|
8.110
|
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr9_+_14135884
|
8.080
|
|
Sesn3
|
sestrin 3
|
|
chr13_+_38243146
|
8.047
|
NM_023842
|
Dsp
|
desmoplakin
|
|
chr4_+_129725083
|
8.035
|
NM_028266
|
Col16a1
|
collagen, type XVI, alpha 1
|
|
chr15_+_78860623
|
7.918
|
|
|
|
|
chr9_-_7932402
|
7.907
|
|
Yap1
|
yes-associated protein 1
|
|
chr14_+_76904316
|
7.730
|
NM_009366
|
Tsc22d1
|
TSC22 domain family, member 1
|
|
chrX_-_131343759
|
7.672
|
NM_001166397
NM_001166398
NM_026139
|
Armcx2
|
armadillo repeat containing, X-linked 2
|
|
chr4_+_138012860
|
7.638
|
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr9_-_83003967
|
7.578
|
|
|
|
|
chr10_+_96945972
|
7.554
|
|
Dcn
|
decorin
|
|
chr2_+_136539202
|
7.474
|
|
Snap25
|
synaptosomal-associated protein 25
|
|
chr14_+_124059330
|
7.473
|
NM_145467
|
Itgbl1
|
integrin, beta-like 1
|
|
chr3_-_126646303
|
7.438
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr14_+_103912557
|
7.431
|
NM_022886
|
Scel
|
sciellin
|
|
chr7_-_149763600
|
7.422
|
|
H19
|
H19 fetal liver mRNA
|
|
chr4_-_117881591
|
7.299
|
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F
|
|
chr15_-_102008126
|
7.298
|
|
|
|
|
chr5_-_137755361
|
7.231
|
|
Trip6
|
thyroid hormone receptor interactor 6
|
|
chr1_-_66991625
|
7.045
|
NM_021285
|
Myl1
|
myosin, light polypeptide 1
|
|
chr15_+_78860043
|
6.914
|
|
H1f0
|
H1 histone family, member 0
|
|
chr5_-_137755406
|
6.906
|
NM_011639
|
Trip6
|
thyroid hormone receptor interactor 6
|
|
chr8_-_74035667
|
6.886
|
NM_032398
|
Plvap
|
plasmalemma vesicle associated protein
|
|
chr3_-_129035406
|
6.878
|
NM_007934
|
Enpep
|
glutamyl aminopeptidase
|
|
chrX_+_139997806
|
6.849
|
NM_008778
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr9_+_118949880
|
6.819
|
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr1_-_172040621
|
6.707
|
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr1_+_45402907
|
6.689
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr14_+_102239673
|
6.679
|
|
Lmo7
|
LIM domain only 7
|
|
chr17_-_34880784
|
6.679
|
NM_009780
|
C4b
|
complement component 4B (Childo blood group)
|
|
chr2_+_20644000
|
6.654
|
|
Etl4
|
enhancer trap locus 4
|
|
chr19_+_30020236
|
6.646
|
NM_001164724
|
Il33
|
interleukin 33
|
|
chr15_+_98462650
|
6.639
|
NM_001044741
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr13_-_29043539
|
6.627
|
|
Sox4
|
SRY-box containing gene 4
|
|
chr11_-_99250663
|
6.621
|
NM_010660
|
Krt10
|
keratin 10
|
|
chr15_-_34608458
|
6.605
|
NM_145469
|
Nipal2
|
NIPA-like domain containing 2
|
|
chr8_-_74150754
|
6.539
|
|
Unc13a
|
unc-13 homolog A (C. elegans)
|
|
chr2_-_140497116
|
6.537
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_+_86578167
|
6.289
|
NM_026414
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chrX_+_38754605
|
6.284
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr8_-_47292973
|
6.259
|
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4
|
|
chrX_+_38754568
|
6.220
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr10_-_36855668
|
6.213
|
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr3_-_27316628
|
6.204
|
|
Fndc3b
|
fibronectin type III domain containing 3B
|
|
chr3_-_84024766
|
6.198
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr6_+_86578550
|
6.119
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr8_-_11199802
|
6.072
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr1_+_108757751
|
6.059
|
NM_009257
|
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5
|
|
chrX_+_38754480
|
6.042
|
NM_016886
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr3_+_67696141
|
5.982
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr1_+_19202046
|
5.973
|
NM_001025305
|
Tcfap2b
|
transcription factor AP-2 beta
|
|
chr2_-_140497183
|
5.966
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr7_-_77511607
|
5.955
|
NM_183261
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_-_140497204
|
5.887
|
NM_001172160
NM_178382
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr17_+_75578120
|
5.870
|
NM_206958
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_97248556
|
5.833
|
NM_001122952
|
Nfia
D130039L10Rik
|
nuclear factor I/A
RIKEN cDNA D130039L10 gene
|
|
chr13_+_23830671
|
5.756
|
NM_015786
|
Hist1h1c
|
histone cluster 1, H1c
|
|
chr8_-_74035629
|
5.748
|
|
Plvap
|
plasmalemma vesicle associated protein
|
|
chr12_+_110800212
|
5.725
|
|
Meg3
|
maternally expressed 3
|
|
chr16_+_45094214
|
5.694
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr16_-_85862579
|
5.687
|
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr7_+_138087002
|
5.641
|
|
Htra1
|
HtrA serine peptidase 1
|
|
chr9_-_86589782
|
5.640
|
NM_008615
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr12_-_81269174
|
5.621
|
|
Actn1
|
actinin, alpha 1
|
|
chr12_-_87419813
|
5.618
|
NM_009368
|
Tgfb3
|
transforming growth factor, beta 3
|
|
chr16_+_35001358
|
5.616
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chrX_+_133242520
|
5.603
|
NM_146236
|
Tceal1
|
transcription elongation factor A (SII)-like 1
|
|
chr1_-_172018982
|
5.579
|
NM_022563
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr12_-_102988596
|
5.572
|
|
|
|
|
chr14_-_21437545
|
5.558
|
NM_030180
|
Usp54
|
ubiquitin specific peptidase 54
|
|
chr17_-_34875879
|
5.555
|
|
C4b
|
complement component 4B (Childo blood group)
|
|
chr7_-_106501642
|
5.506
|
NM_001111043
NM_001111044
NM_009825
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr15_+_98465328
|
5.485
|
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr8_+_81552335
|
5.476
|
NM_178267
|
Zfp827
|
zinc finger protein 827
|
|
chr15_-_99650502
|
5.432
|
NM_023063
|
Lima1
|
LIM domain and actin binding 1
|
|
chr10_+_56097078
|
5.419
|
NM_010288
|
Gja1
|
gap junction protein, alpha 1
|
|
chr1_+_155756310
|
5.394
|
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase)
|
|
chr16_+_4639921
|
5.369
|
NM_139307
|
Vasn
|
vasorin
|
|
chr9_-_91260597
|
5.342
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr7_+_52965052
|
5.280
|
|
Dbp
|
D site albumin promoter binding protein
|
|
chr16_+_45094163
|
5.280
|
NM_026439
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr1_+_24258538
|
5.279
|
|
Col9a1
|
collagen, type IX, alpha 1
|
|
chr19_+_10525829
|
5.260
|
|
|
|
|
chr4_-_60889249
|
5.251
|
NM_001134674
|
Mup13
Mup19
Mup1
|
major urinary protein 13
major urinary protein 19
major urinary protein 1
|
|
chr2_-_165709697
|
5.248
|
|
Zmynd8
|
zinc finger, MYND-type containing 8
|
|
chr4_-_60594469
|
5.234
|
|
Mup1
|
major urinary protein 1
|
|
chr15_-_34608600
|
5.228
|
|
Nipal2
|
NIPA-like domain containing 2
|
|
chr4_-_73436505
|
5.217
|
NM_001017427
|
Rasef
|
RAS and EF hand domain containing
|
|
chr6_-_38238095
|
5.151
|
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like
|
|
chr5_-_104058250
|
5.142
|
NM_029415
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr3_-_113277601
|
5.141
|
NM_007446
|
Amy1
|
amylase 1, salivary
|
|
chr19_+_38598686
|
5.127
|
NM_019588
|
Plce1
|
phospholipase C, epsilon 1
|
|
chr15_+_10882086
|
5.093
|
NM_030888
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3
|
|
chr1_-_82232392
|
5.052
|
|
Irs1
|
insulin receptor substrate 1
|
|
chr17_-_90436488
|
5.049
|
|
Nrxn1
|
neurexin I
|
|
chr18_+_65739883
|
5.036
|
NM_207255
|
Zfp532
|
zinc finger protein 532
|
|
chr3_-_83967952
|
5.023
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr14_+_66285263
|
5.021
|
|
Scara5
|
scavenger receptor class A, member 5 (putative)
|
|
chr4_-_60594496
|
4.982
|
|
Mup1
|
major urinary protein 1
|
|
chr3_+_133903056
|
4.977
|
NM_001004367
|
Cxxc4
|
CXXC finger 4
|
|
chr7_-_31861665
|
4.959
|
NM_008557
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3
|
|
chr6_-_148937854
|
4.936
|
|
Dennd5b
|
DENN/MADD domain containing 5B
|
|
chr16_+_96422076
|
4.900
|
NM_015825
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr18_+_37678006
|
4.899
|
NM_053147
|
Pcdhb22
|
protocadherin beta 22
|
|
chr11_+_102466556
|
4.896
|
|
Fzd2
|
frizzled homolog 2 (Drosophila)
|
|
chr11_+_100277007
|
4.883
|
NM_001163481
NM_010221
|
Fkbp10
|
FK506 binding protein 10
|
|
chrX_+_162782484
|
4.870
|
NM_001177955
NM_001177956
NM_001177957
NM_001177958
NM_001177959
NM_001177960
|
Gpm6b
|
glycoprotein m6b
|
|
chr3_+_85888818
|
4.822
|
|
Sh3d19
|
SH3 domain protein D19
|
|
chr8_+_56040211
|
4.774
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr16_-_44558929
|
4.765
|
NM_172506
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein
|
|
chr16_+_45093784
|
4.762
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr3_-_122241909
|
4.729
|
|
Fnbp1l
|
formin binding protein 1-like
|
|
chr2_+_169458802
|
4.684
|
|
Tshz2
|
teashirt zinc finger family member 2
|
|
chr19_+_26822892
|
4.662
|
NM_026003
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr18_+_80243633
|
4.621
|
NM_053117
|
Pard6g
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr1_-_66991580
|
4.604
|
|
Myl1
|
myosin, light polypeptide 1
|
|
chr3_-_88285172
|
4.592
|
|
|
|
|
chr13_+_43218207
|
4.575
|
NM_001005748
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr11_+_98537516
|
4.544
|
|
Gsdma
|
gasdermin A
|
|
chr10_-_36855292
|
4.543
|
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr8_-_67071497
|
4.524
|
|
|
|
|
chr1_-_132586370
|
4.492
|
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr11_+_96711152
|
4.484
|
NM_019877
|
Copz2
|
coatomer protein complex, subunit zeta 2
|
|
chr7_-_4116300
|
4.479
|
NM_021454
|
Cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chrX_+_133357268
|
4.448
|
NM_011123
|
Plp1
|
proteolipid protein (myelin) 1
|
|
chr4_+_102432943
|
4.418
|
NM_144906
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr4_+_135700717
|
4.370
|
|
Id3
|
inhibitor of DNA binding 3
|
|
chr7_-_149763980
|
4.353
|
|
H19
|
H19 fetal liver mRNA
|
|
chr11_+_98913612
|
4.344
|
|
|
|
|
chrX_-_137908516
|
4.287
|
|
Col4a6
|
collagen, type IV, alpha 6
|
|
chr8_+_123640069
|
4.272
|
NM_013519
|
Foxc2
|
forkhead box C2
|