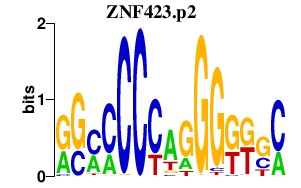
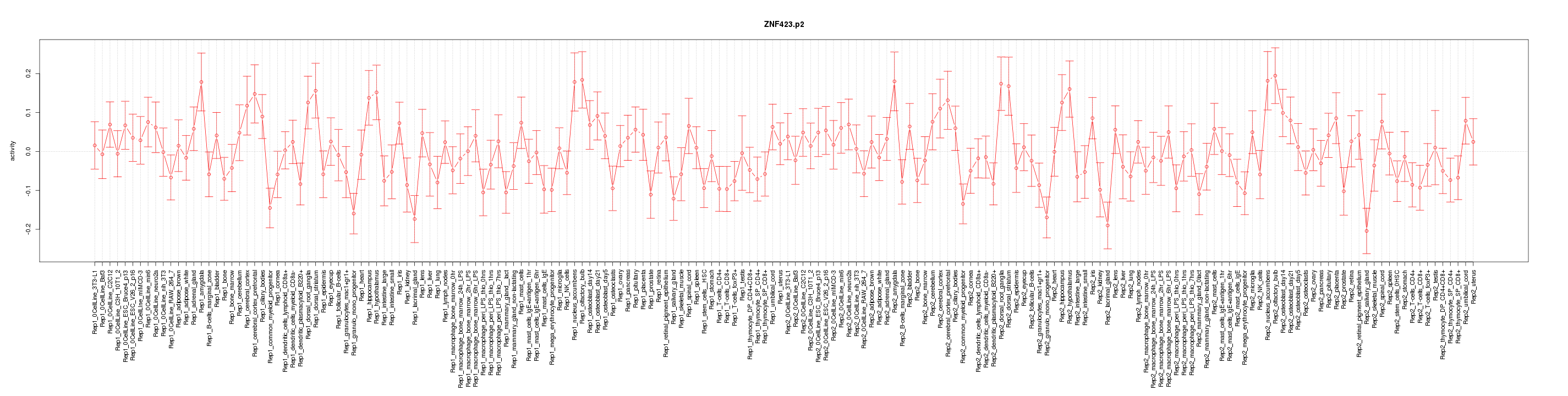
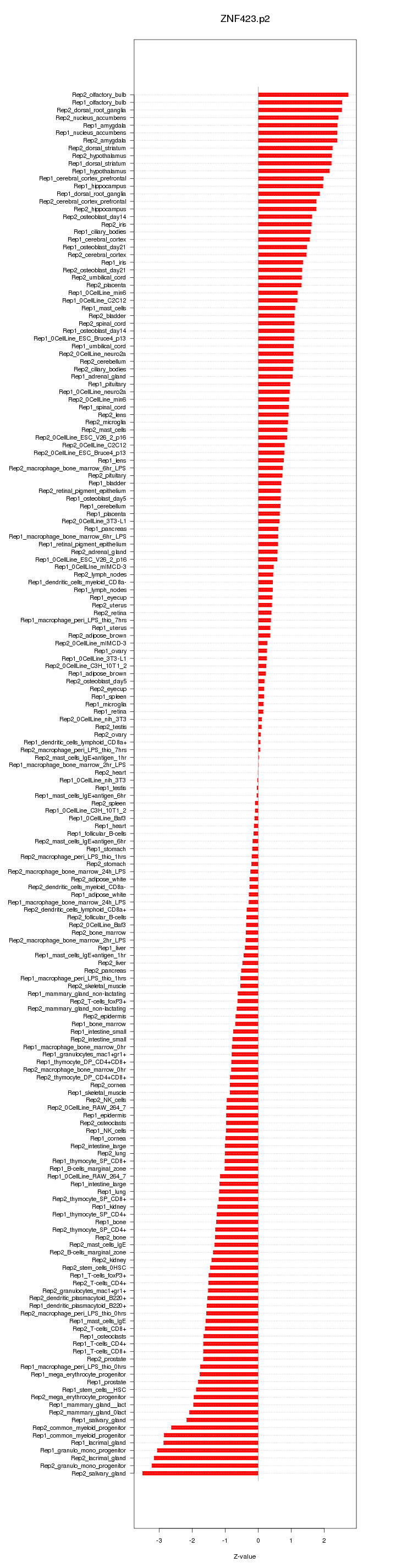
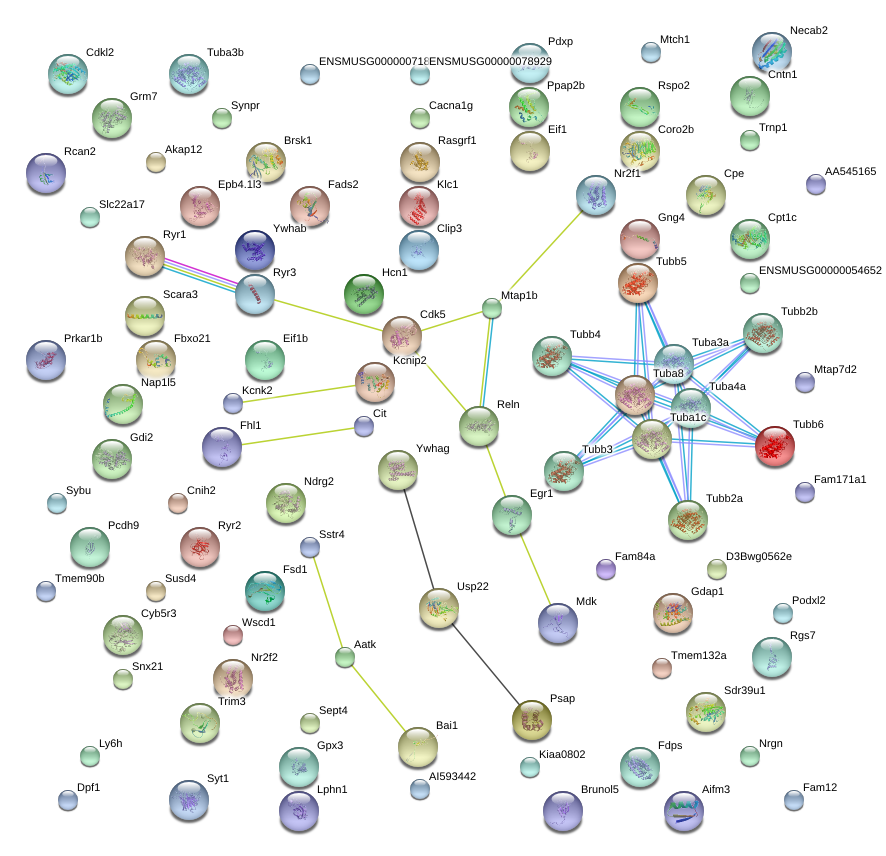
|
chr17_+_69506165
|
27.747
|
|
Epb4.1l3
|
erythrocyte protein band 4.1-like 3
|
|
chr6_-_58857000
|
21.812
|
NM_021432
|
Nap1l5
|
nucleosome assembly protein 1-like 5
|
|
chr17_+_69506083
|
20.376
|
NM_013813
|
Epb4.1l3
|
erythrocyte protein band 4.1-like 3
|
|
chr8_+_98201984
|
17.675
|
|
|
|
|
chr8_-_67171481
|
16.870
|
|
Cpe
|
carboxypeptidase E
|
|
chr11_+_87391881
|
16.860
|
|
Sept4
|
septin 4
|
|
chr9_+_106366204
|
15.513
|
|
|
|
|
chr8_-_67171805
|
15.277
|
NM_013494
|
Cpe
|
carboxypeptidase E
|
|
chr5_-_136410393
|
15.009
|
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr1_+_184695026
|
14.973
|
NM_144796
|
Susd4
|
sushi domain containing 4
|
|
chr10_-_108447996
|
14.729
|
|
Syt1
|
synaptotagmin I
|
|
chr19_-_10175969
|
13.424
|
|
Fads2
|
fatty acid desaturase 2
|
|
chr5_-_136410216
|
13.327
|
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr7_+_31076771
|
13.294
|
NM_001081114
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr6_+_110595586
|
12.588
|
NM_177328
|
Grm7
|
glutamate receptor, metabotropic 7
|
|
chr8_+_125935470
|
12.532
|
|
Tubb3
|
tubulin, beta 3
|
|
chr10_-_80945445
|
12.505
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr13_-_100286521
|
12.483
|
NM_008634
|
Mtap1b
|
microtubule-associated protein 1B
|
|
chr7_-_52230091
|
12.397
|
|
Cpt1c
|
carnitine palmitoyltransferase 1c
|
|
chr13_-_100286446
|
12.354
|
|
Mtap1b
|
microtubule-associated protein 1B
|
|
chr4_-_133053867
|
12.177
|
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr10_-_108448003
|
12.168
|
|
Syt1
|
synaptotagmin I
|
|
chr9_-_52487410
|
11.864
|
NM_178906
|
AI593442
|
expressed sequence AI593442
|
|
chr6_-_88824037
|
11.739
|
NM_176973
|
Podxl2
|
podocalyxin-like 2
|
|
chr11_-_119878383
|
11.557
|
|
Aatk
|
apoptosis-associated tyrosine kinase
|
|
chr12_+_112997057
|
11.419
|
NM_001025360
NM_001081959
NM_008450
|
Klc1
|
kinesin light chain 1
|
|
chr14_+_14117313
|
11.342
|
|
Synpr
|
synaptoporin
|
|
chr18_+_40416615
|
11.292
|
|
2900055J20Rik
|
RIKEN cDNA 2900055J20 gene
|
|
chr9_-_37360141
|
11.219
|
NM_022029
|
Nrgn
|
neurogranin
|
|
chr8_+_121970569
|
11.176
|
NM_054095
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr14_+_14117292
|
10.909
|
NM_028052
|
Synpr
|
synaptoporin
|
|
chr9_+_120401723
|
10.685
|
NM_026892
|
Eif1b
|
eukaryotic translation initiation factor 1B
|
|
chr2_+_3035692
|
10.502
|
|
Fam171a1
|
family with sequence similarity 171, member A1
|
|
chr19_-_5098486
|
10.465
|
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr19_-_10944196
|
10.454
|
NM_133804
|
Tmem132a
|
transmembrane protein 132A
|
|
chr7_-_52230179
|
10.374
|
|
Cpt1c
|
carnitine palmitoyltransferase 1c
|
|
chr15_-_44619541
|
10.256
|
NM_001032727
NM_176998
|
Sybu
|
syntabulin (syntaxin-interacting)
|
|
chr15_+_78744802
|
10.163
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr19_-_5098498
|
10.152
|
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr14_-_55531726
|
10.039
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr10_-_61573705
|
10.020
|
NM_027251
|
2010107G23Rik
|
RIKEN cDNA 2010107G23 gene
|
|
chr7_-_112781902
|
9.956
|
NM_018880
|
Trim3
|
tripartite motif-containing 3
|
|
chr19_-_10175990
|
9.950
|
NM_019699
|
Fads2
|
fatty acid desaturase 2
|
|
chr8_+_125935441
|
9.927
|
NM_023279
|
Tubb3
|
tubulin, beta 3
|
|
chr14_-_66539632
|
9.922
|
|
Scara3
|
scavenger receptor class A, member 3
|
|
chr3_-_117063090
|
9.858
|
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr9_-_62384483
|
9.841
|
|
Coro2b
|
coronin, actin binding protein, 2B
|
|
chr9_+_89804612
|
9.706
|
NM_001039655
NM_011245
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1
|
|
chr10_-_80945379
|
9.640
|
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr11_-_60988490
|
9.458
|
NM_001004143
|
Usp22
|
ubiquitin specific peptidase 22
|
|
chr5_-_21850490
|
9.446
|
NM_011261
|
Reln
|
reelin
|
|
chr4_+_104830022
|
9.401
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr19_-_5098381
|
9.398
|
NM_009920
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr11_-_60988419
|
9.360
|
|
Usp22
|
ubiquitin specific peptidase 22
|
|
chr12_-_14158843
|
9.269
|
NM_029007
|
Fam84a
|
family with sequence similarity 84, member A
|
|
chr7_-_17501314
|
9.145
|
|
|
|
|
chr1_+_17135421
|
9.119
|
NM_010267
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1
|
|
chr5_+_116295259
|
9.104
|
|
Cit
|
citron
|
|
chr15_+_91881593
|
9.091
|
NM_001159648
|
Cntn1
|
contactin 1
|
|
chr2_+_149656518
|
8.984
|
NM_001085521
|
Tmem90b
|
transmembrane protein 90B
|
|
chr7_-_52230117
|
8.792
|
NM_153679
|
Cpt1c
|
carnitine palmitoyltransferase 1c
|
|
chr13_+_13876570
|
8.747
|
|
Gng4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr14_-_94289847
|
8.618
|
|
Pcdh9
|
protocadherin 9
|
|
chr17_+_56125891
|
8.613
|
NM_183178
|
Fsd1
|
fibronectin type 3 and SPRY domain-containing protein
|
|
chr9_+_120401824
|
8.477
|
|
Eif1b
|
eukaryotic translation initiation factor 1B
|
|
chr10_-_108447894
|
8.386
|
|
Syt1
|
synaptotagmin I
|
|
chr15_-_75397601
|
8.370
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr15_-_83002561
|
8.312
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr5_-_23929349
|
8.255
|
|
Cdk5
|
cyclin-dependent kinase 5
|
|
chr16_+_17489743
|
8.139
|
NM_175178
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3
|
|
chr11_+_54716354
|
8.081
|
NM_001083929
NM_008161
|
Gpx3
|
glutathione peroxidase 3
|
|
chrX_+_53985133
|
8.049
|
NM_001077361
|
Fhl1
|
four and a half LIM domains 1
|
|
chr12_+_112997194
|
7.855
|
|
Klc1
|
kinesin light chain 1
|
|
chrX_+_53984926
|
7.786
|
NM_010211
|
Fhl1
|
four and a half LIM domains 1
|
|
chr6_+_145564482
|
7.765
|
NM_009449
|
Tuba3b
|
tubulin, alpha 3B
|
|
chr17_+_43938789
|
7.744
|
NM_207649
|
Rcan2
|
regulator of calcineurin 2
|
|
chr15_+_74394184
|
7.705
|
|
Bai1
|
brain-specific angiogenesis inhibitor 1
|
|
chr15_-_83002624
|
7.590
|
NM_029787
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr5_-_23929347
|
7.534
|
NM_007668
|
Cdk5
|
cyclin-dependent kinase 5
|
|
chr8_+_86423968
|
7.527
|
NM_181039
|
Lphn1
|
latrophilin 1
|
|
chr13_-_78337844
|
7.499
|
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr10_-_6080145
|
7.475
|
NM_031185
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12
|
|
chr11_+_54716439
|
7.314
|
|
Gpx3
|
glutathione peroxidase 3
|
|
chr13_+_118391126
|
7.309
|
NM_010408
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1
|
|
chr15_-_83002612
|
7.227
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr2_-_91771919
|
7.189
|
NM_001012336
|
Mdk
|
midkine
|
|
chr5_-_139605617
|
7.148
|
NM_008923
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta
|
|
chr7_+_4642529
|
7.103
|
NM_001003920
NM_001168572
|
Brsk1
|
BR serine/threonine kinase 1
|
|
chr5_-_136410488
|
7.087
|
NM_018871
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr15_-_43002356
|
7.019
|
NM_172815
|
Rspo2
|
R-spondin 2 homolog (Xenopus laevis)
|
|
chr18_+_35020867
|
6.691
|
|
Egr1
|
early growth response 1
|
|
chr9_+_89804723
|
6.671
|
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1
|
|
chr10_+_59763520
|
6.653
|
|
Psap
|
prosaposin
|
|
chr7_+_31076852
|
6.609
|
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr13_-_12199109
|
6.584
|
NM_023868
|
Ryr2
|
ryanodine receptor 2, cardiac
|
|
chr14_-_56519012
|
6.569
|
NM_001082975
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr2_+_163820857
|
6.540
|
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr2_-_91771882
|
6.479
|
|
Mdk
|
midkine
|
|
chr2_-_91771836
|
6.476
|
NM_001012335
|
Mdk
|
midkine
|
|
chr18_+_35020859
|
6.426
|
NM_007913
|
Egr1
|
early growth response 1
|
|
chr3_-_88905775
|
6.381
|
NM_134469
|
Fdps
|
farnesyl diphosphate synthetase
|
|
chr1_-_191167582
|
6.343
|
NM_010607
|
Kcnk2
|
potassium channel, subfamily K, member 2
|
|
chr11_-_94335350
|
6.339
|
NM_001112813
NM_001177888
NM_001177890
NM_009783
|
Cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr7_+_30093030
|
6.317
|
|
Dpf1
|
D4, zinc and double PHD fingers family 1
|
|
chr13_+_3537514
|
6.309
|
|
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2
|
|
chr11_+_71564204
|
6.298
|
NM_177618
|
Wscd1
|
WSC domain containing 1
|
|
chr2_+_148221069
|
6.283
|
|
Sstr4
|
somatostatin receptor 4
|
|
chr17_+_43938887
|
6.281
|
|
Rcan2
|
regulator of calcineurin 2
|
|
chr15_+_74346835
|
6.262
|
|
Bai1
|
brain-specific angiogenesis inhibitor 1
|
|
chrX_+_155852641
|
6.256
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr2_+_164611444
|
6.230
|
NM_133924
|
Snx21
|
sorting nexin family member 21
|
|
chr5_+_118426802
|
6.211
|
|
Fbxo21
|
F-box protein 21
|
|
chr15_+_74346853
|
6.153
|
|
Bai1
|
brain-specific angiogenesis inhibitor 1
|
|
chr8_+_121970660
|
6.148
|
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr17_-_66799086
|
5.882
|
NM_001114098
NM_172963
|
1110012J17Rik
|
RIKEN cDNA 1110012J17 gene
|
|
chr19_-_45890238
|
5.853
|
NM_030716
NM_145703
NM_145704
|
Kcnip2
|
Kv channel-interacting protein 2
|
|
chr14_-_52533114
|
5.800
|
NM_001145959
NM_013864
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr1_-_177422520
|
5.721
|
|
Rgs7
|
regulator of G protein signaling 7
|
|
chr5_-_92471976
|
5.701
|
NM_177270
NM_016912
|
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr9_+_103014463
|
5.526
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr7_-_52124950
|
5.519
|
|
Ptov1
|
prostate tumor over expressed gene 1
|
|
chr9_+_20673069
|
5.473
|
NM_175687
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene
|
|
chr8_+_11312828
|
5.463
|
NM_009932
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr8_-_11312686
|
5.458
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr7_+_64846558
|
5.404
|
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr17_-_25570566
|
5.363
|
|
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr6_+_48487648
|
5.357
|
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr7_+_64846528
|
5.326
|
NM_008071
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr2_+_158436493
|
5.305
|
NM_009508
|
Slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr4_+_129190824
|
5.243
|
NM_010807
|
Marcksl1
|
MARCKS-like 1
|
|
chr2_+_121692805
|
5.239
|
|
Casc4
|
cancer susceptibility candidate 4
|
|
chr10_-_108447674
|
5.165
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chrX_+_155852439
|
5.137
|
NM_001081124
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr2_-_30214724
|
5.106
|
NM_139302
|
Sh3glb2
|
SH3-domain GRB2-like endophilin B2
|
|
chr5_-_23929280
|
5.102
|
|
Cdk5
|
cyclin-dependent kinase 5
|
|
chr11_+_29593657
|
5.100
|
NM_194051
|
Rtn4
|
reticulon 4
|
|
chr8_+_121971001
|
5.024
|
|
Necab2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr1_-_164408008
|
4.986
|
|
Dnm3
|
dynamin 3
|
|
chr7_+_25266986
|
4.961
|
NM_153112
|
Cadm4
|
cell adhesion molecule 4
|
|
chr13_-_19711409
|
4.950
|
NM_134065
|
Epdr1
|
ependymin related protein 1 (zebrafish)
|
|
chr13_-_69877709
|
4.868
|
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr9_+_45646611
|
4.855
|
NM_001145947
NM_011792
|
Bace1
|
beta-site APP cleaving enzyme 1
|
|
chr13_-_78338131
|
4.791
|
NM_010151
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr16_+_58408609
|
4.734
|
NM_028523
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr11_-_69373555
|
4.732
|
|
Efnb3
|
ephrin B3
|
|
chr15_+_103333554
|
4.637
|
NM_008800
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr9_+_20673178
|
4.629
|
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene
|
|
chr11_-_102680999
|
4.602
|
NM_001159383
|
Gjc1
|
gap junction protein, gamma 1
|
|
chr11_+_98629709
|
4.538
|
|
|
|
|
chr14_-_52532988
|
4.514
|
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr8_-_75876420
|
4.504
|
NM_010687
|
Large
|
like-glycosyltransferase
|
|
chr5_+_67652034
|
4.416
|
|
Tmem33
|
transmembrane protein 33
|
|
chr4_-_58566053
|
4.334
|
NM_172989
|
Lpar1
|
lysophosphatidic acid receptor 1
|
|
chr14_-_52533039
|
4.316
|
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr2_+_38194775
|
4.294
|
|
Lhx2
|
LIM homeobox protein 2
|
|
chr4_+_43681807
|
4.273
|
NM_001085508
|
Tmem8b
|
transmembrane protein 8B
|
|
chr8_-_114267646
|
4.215
|
NM_009954
|
Bcar1
|
breast cancer anti-estrogen resistance 1
|
|
chr16_+_6349108
|
4.206
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_+_118426792
|
4.204
|
|
Fbxo21
|
F-box protein 21
|
|
chr12_-_60162382
|
4.189
|
NM_030057
|
Trappc6b
|
trafficking protein particle complex 6B
|
|
chr11_-_87800086
|
4.163
|
NM_001168472
|
Dynll2
|
dynein light chain LC8-type 2
|
|
chr15_-_100382354
|
4.148
|
NM_033476
|
Tcfcp2
|
transcription factor CP2
|
|
chr15_+_74346624
|
4.091
|
NM_174991
|
Bai1
|
brain-specific angiogenesis inhibitor 1
|
|
chr10_+_13686174
|
4.088
|
NM_010437
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr19_+_10116079
|
4.072
|
|
Fads3
|
fatty acid desaturase 3
|
|
chr13_+_41345120
|
4.066
|
NM_001135577
|
BC024659
|
cDNA sequence BC024659
|
|
chr17_-_57199207
|
4.045
|
NM_025877
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr7_-_119301920
|
4.036
|
|
Dkk3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr8_-_74195650
|
4.035
|
NM_001029873
|
Unc13a
|
unc-13 homolog A (C. elegans)
|
|
chr2_+_119625463
|
4.019
|
|
Tyro3
|
TYRO3 protein tyrosine kinase 3
|
|
chr3_+_96404978
|
4.019
|
NM_001163170
|
Lix1l
|
Lix1-like
|
|
chr3_-_87804062
|
4.012
|
|
Bcan
|
brevican
|
|
chr6_+_72047563
|
4.007
|
NM_001035228
NM_011375
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chrX_+_97963061
|
3.925
|
NM_001177778
NM_016747
|
Dlg3
|
discs, large homolog 3 (Drosophila)
|
|
chr15_+_26238802
|
3.898
|
NM_177597
|
March11
|
membrane-associated ring finger (C3HC4) 11
|
|
chr5_+_22252006
|
3.892
|
NM_001081231
NM_029990
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3
|
|
chr14_-_52935887
|
3.890
|
|
Sall2
|
sal-like 2 (Drosophila)
|
|
chr13_+_19040215
|
3.882
|
NM_175007
|
Amph
|
amphiphysin
|
|
chr5_+_116295664
|
3.880
|
NM_007708
|
Cit
|
citron
|
|
chr9_-_108499855
|
3.875
|
NM_028944
|
P4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr4_+_42930076
|
3.824
|
NM_172690
|
N28178
|
expressed sequence N28178
|
|
chr17_+_43938905
|
3.760
|
|
Rcan2
|
regulator of calcineurin 2
|
|
chr12_-_60162456
|
3.725
|
|
Trappc6b
|
trafficking protein particle complex 6B
|
|
chr13_-_54712629
|
3.712
|
NM_028870
|
Cltb
|
clathrin, light polypeptide (Lcb)
|
|
chr15_+_103333440
|
3.708
|
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr5_+_88994019
|
3.644
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr11_+_71564481
|
3.622
|
|
Wscd1
|
WSC domain containing 1
|
|
chr5_+_118426774
|
3.615
|
NM_145564
|
Fbxo21
|
F-box protein 21
|
|
chr1_+_94830952
|
3.610
|
NM_011796
|
Capn10
|
calpain 10
|
|
chr1_-_172519932
|
3.599
|
NM_001109985
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr12_+_79327673
|
3.566
|
|
Gphn
|
gephyrin
|
|
chr10_+_75398317
|
3.555
|
NM_175329
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr8_-_11312729
|
3.547
|
NM_009931
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr2_+_121692687
|
3.543
|
NM_177054
NM_199038
|
Casc4
|
cancer susceptibility candidate 4
|
|
chr16_+_17276720
|
3.539
|
|
Tmem191c
|
transmembrane protein 191C
|
|
chr11_-_69373707
|
3.533
|
NM_007911
|
Efnb3
|
ephrin B3
|
|
chr5_-_143084677
|
3.513
|
NM_175217
|
Mmd2
|
monocyte to macrophage differentiation-associated 2
|
|
chr1_-_175297624
|
3.504
|
|
Cadm3
|
cell adhesion molecule 3
|
|
chr1_+_66515003
|
3.488
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr1_-_177422630
|
3.481
|
NM_001199003
NM_011880
|
Rgs7
|
regulator of G protein signaling 7
|