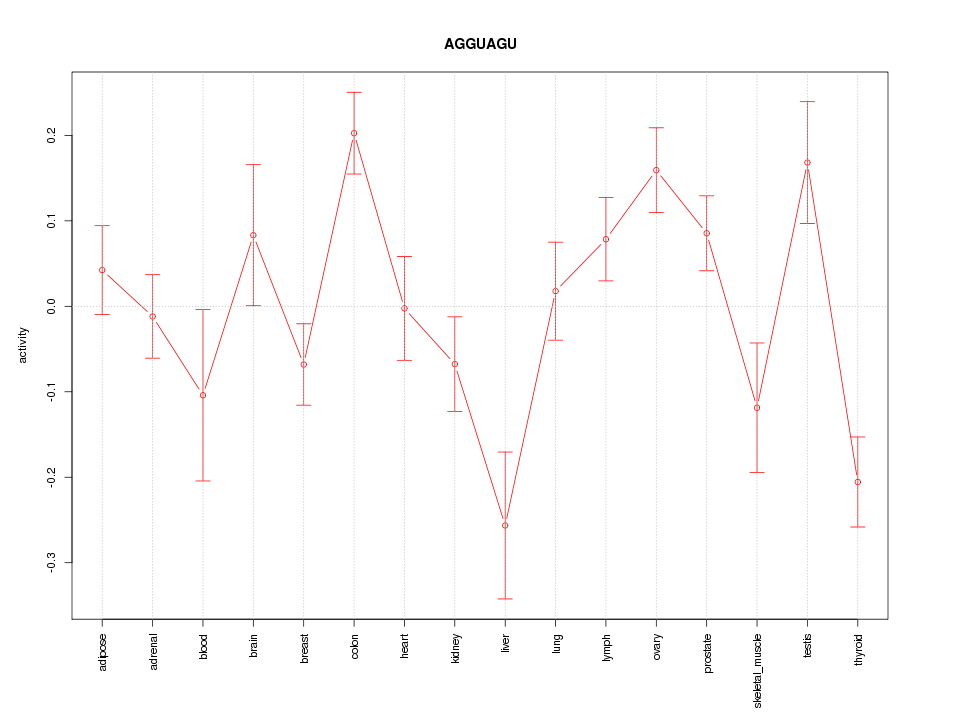
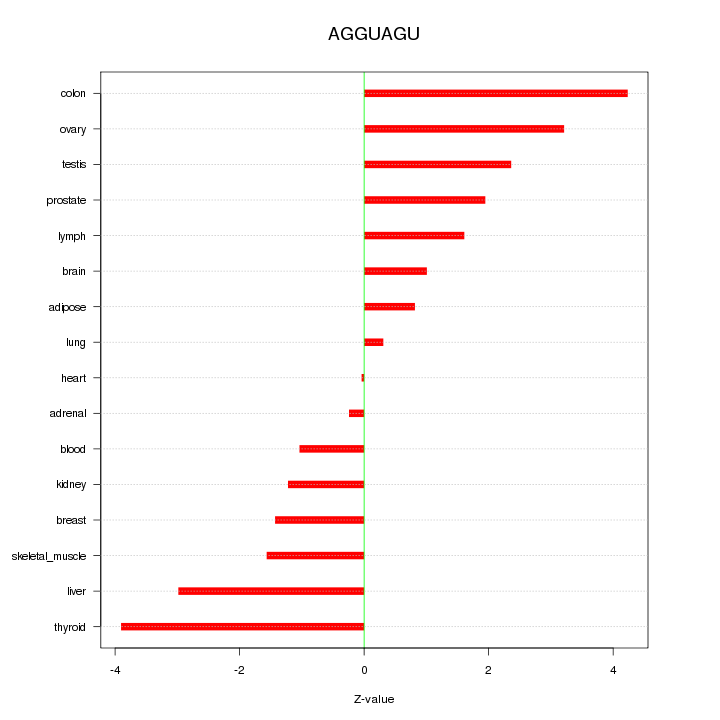
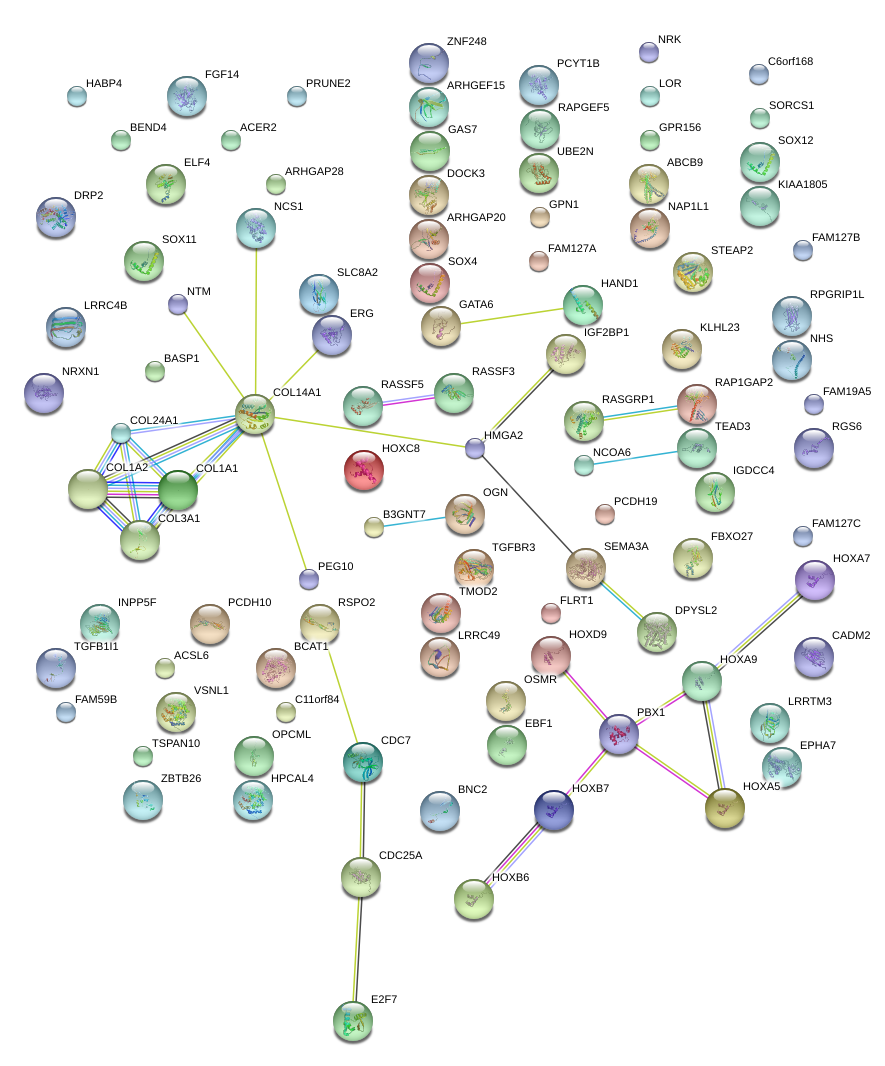
|
chr6_-_94129059
|
5.269
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr22_+_48885255
|
4.602
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr7_-_27196163
|
4.315
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr22_+_48972117
|
3.860
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chrX_+_105066531
|
3.849
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr8_-_109095836
|
3.843
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chr9_-_16870719
|
3.836
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr19_-_51071279
|
3.515
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chrX_-_99665270
|
3.436
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr11_+_63580922
|
3.410
|
NM_138471
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr17_-_46688362
|
3.378
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr17_-_10101854
|
3.318
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr4_-_42154894
|
3.217
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr18_+_6834431
|
3.193
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr5_-_153857820
|
3.166
|
NM_004821
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr19_-_47975244
|
3.154
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr15_+_52043738
|
3.120
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr12_+_54402795
|
3.110
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr17_-_9929567
|
2.855
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr7_+_94023872
|
2.837
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr12_-_123451031
|
2.822
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr9_+_19408924
|
2.800
|
NM_001010887
|
ACER2
|
alkaline ceramidase 2
|
|
chr12_-_25102281
|
2.799
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chrX_-_24665454
|
2.717
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr3_-_119963141
|
2.713
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr7_+_94285620
|
2.697
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr11_+_63871240
|
2.656
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr9_+_132962871
|
2.646
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr6_-_99797530
|
2.645
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr5_+_38845962
|
2.641
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr15_-_65715385
|
2.613
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr11_-_132812870
|
2.573
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr9_+_132934685
|
2.481
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chrX_-_24690740
|
2.444
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr7_+_94285676
|
2.431
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr7_+_89840999
|
2.428
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr2_+_232260334
|
2.408
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr17_-_9940004
|
2.323
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_48278987
|
2.239
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_-_51259536
|
2.190
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr12_-_25055228
|
2.029
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr3_-_48229800
|
2.024
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr10_+_68685791
|
2.014
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr2_-_50574891
|
1.991
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr17_-_9862765
|
1.953
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr17_+_2699731
|
1.895
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr17_-_46682333
|
1.806
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr12_-_25055966
|
1.735
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_+_65004191
|
1.701
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr7_-_83824216
|
1.627
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr11_-_133402227
|
1.567
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr9_-_79520878
|
1.563
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr3_+_50712671
|
1.555
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr2_+_5832778
|
1.500
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chrX_+_100474932
|
1.490
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr9_+_99212379
|
1.470
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr17_+_8214139
|
1.453
|
NM_025014
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr12_-_76478737
|
1.449
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr17_+_47074749
|
1.437
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr8_+_121137297
|
1.387
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr1_+_164528586
|
1.358
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_+_153232178
|
1.296
|
NM_000427
|
LOR
|
loricrin
|
|
chr5_-_131347518
|
1.269
|
NM_001205248
NM_001205251
NM_001205250
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr3_+_85008031
|
1.266
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr9_+_123850573
|
1.248
|
NM_007018
|
CNTRL
|
centriolin
|
|
chr15_+_71184781
|
1.244
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr12_+_66218141
|
1.219
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr7_-_27205148
|
1.193
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr10_-_38146221
|
1.177
|
NM_021045
|
ZNF248
|
zinc finger protein 248
|
|
chr2_+_189839078
|
1.172
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr13_-_102568994
|
1.147
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr1_-_92371558
|
1.123
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_170590241
|
1.111
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr3_+_85775631
|
1.100
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr15_-_38856830
|
1.098
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr16_+_31484497
|
1.096
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr2_+_17721806
|
1.057
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chrX_-_134156488
|
1.055
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr20_+_306205
|
1.053
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr7_-_27183225
|
1.051
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr14_+_72399785
|
1.024
|
NM_001204416
NM_001204417
NM_001204418
NM_001204419
NM_001204420
NM_001204421
NM_001204422
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr17_+_8213555
|
1.016
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr6_-_35464716
|
0.974
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr1_-_86622107
|
0.956
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chr18_+_19749403
|
0.949
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr14_+_72398816
|
0.941
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chrX_+_17653412
|
0.920
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr13_-_103054027
|
0.919
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr1_+_91966365
|
0.916
|
NM_001134419
NM_003503
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr16_-_53737713
|
0.894
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr2_+_26395959
|
0.882
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr2_+_27805892
|
0.881
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr16_+_31483066
|
0.869
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr9_-_125693778
|
0.868
|
NM_020924
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
|
chrX_-_129244471
|
0.868
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr7_-_22396511
|
0.867
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr11_-_110583120
|
0.864
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr9_-_95166827
|
0.849
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr5_-_158526698
|
0.847
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr19_-_39523132
|
0.832
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr21_-_39870303
|
0.822
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_77459198
|
0.822
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr5_+_17217474
|
0.783
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr10_-_108924287
|
0.780
|
NM_001013031
NM_001206569
NM_001206570
NM_001206571
NM_001206572
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr11_+_44785975
|
0.741
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr21_-_40033617
|
0.739
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_+_91966653
|
0.739
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chrX_-_129244652
|
0.737
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chrX_-_140271293
|
0.693
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr5_+_172068188
|
0.691
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr1_-_92351613
|
0.691
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chrX_+_17393537
|
0.690
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr8_-_82754427
|
0.687
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr22_+_39883228
|
0.684
|
NM_001098270
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr1_+_50513685
|
0.680
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr9_-_140196702
|
0.671
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr8_+_28351694
|
0.664
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr19_+_47104490
|
0.636
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr6_-_84419108
|
0.633
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr14_+_72399149
|
0.631
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr2_+_26403584
|
0.620
|
NM_001191033
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr4_-_122854192
|
0.618
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chrX_+_134166318
|
0.600
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr4_+_174089889
|
0.597
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr2_+_172378856
|
0.586
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr5_+_153825497
|
0.570
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr6_-_42419782
|
0.563
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr12_-_105478218
|
0.544
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr9_+_6413139
|
0.535
|
NM_152896
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr5_-_74326723
|
0.535
|
NM_016591
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chr22_+_39853316
|
0.529
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr3_+_89156673
|
0.525
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr5_-_131347249
|
0.520
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr6_-_84418705
|
0.520
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr7_-_45960738
|
0.505
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr8_-_81787015
|
0.496
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr12_-_57472573
|
0.492
|
NM_001130963
NM_015257
|
TMEM194A
|
transmembrane protein 194A
|
|
chr13_-_33760215
|
0.486
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_-_90758126
|
0.484
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_-_60780404
|
0.476
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chrX_-_74145280
|
0.464
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr8_+_1449531
|
0.464
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr11_-_798196
|
0.463
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr5_+_148724976
|
0.458
|
NM_152407
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr1_-_179112178
|
0.451
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr1_+_36023389
|
0.442
|
NM_001014839
NM_001014841
NM_014284
|
NCDN
|
neurochondrin
|
|
chr12_+_27933162
|
0.442
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chrX_-_83757398
|
0.435
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr8_+_22102618
|
0.427
|
NM_001722
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|
|
chr12_+_4382882
|
0.425
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr20_-_31071187
|
0.418
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr1_+_50571963
|
0.417
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr9_-_99801365
|
0.414
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr2_-_25194772
|
0.412
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr14_-_57735614
|
0.412
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr10_+_72238529
|
0.408
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chr9_-_77567735
|
0.407
|
NM_017998
|
C9orf40
|
chromosome 9 open reading frame 40
|
|
chr14_+_33408458
|
0.400
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr7_-_559479
|
0.400
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr7_-_138665853
|
0.398
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr4_-_90759446
|
0.393
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_-_175499275
|
0.376
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175547471
|
0.372
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_-_122872908
|
0.361
|
NM_001130698
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr9_+_133710746
|
0.344
|
NM_005157
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr6_-_33267128
|
0.344
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr1_+_113615787
|
0.344
|
NM_014813
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr12_+_123868608
|
0.335
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr3_-_28390506
|
0.334
|
NM_001134432
NM_001134433
NM_022461
|
AZI2
|
5-azacytidine induced 2
|
|
chr3_-_150264280
|
0.333
|
NM_014445
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr3_-_178789597
|
0.327
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr1_+_50574589
|
0.327
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr8_-_67525426
|
0.321
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr6_+_134274247
|
0.313
|
NM_004865
|
TBPL1
|
TBP-like 1
|
|
chr16_-_30621628
|
0.309
|
NM_138447
|
ZNF689
|
zinc finger protein 689
|
|
chr20_-_656822
|
0.307
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr1_-_17765052
|
0.301
|
NM_001136204
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr17_+_28268622
|
0.296
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr3_+_49208979
|
0.285
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr1_+_99729847
|
0.283
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chrX_+_47696281
|
0.282
|
NM_007137
|
ZNF81
|
zinc finger protein 81
|
|
chr4_-_52904424
|
0.280
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr3_+_61547179
|
0.272
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr12_-_93323088
|
0.271
|
NM_003566
|
EEA1
|
early endosome antigen 1
|
|
chr1_+_215256559
|
0.268
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr20_+_30598244
|
0.266
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr12_-_59314069
|
0.265
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr20_+_32077904
|
0.264
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr6_-_134373618
|
0.262
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr7_-_98741681
|
0.255
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr18_+_55816546
|
0.253
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_+_91003238
|
0.248
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr9_-_99801924
|
0.247
|
NM_001201575
|
CTSL2
|
cathepsin L2
|
|
chrX_+_2746810
|
0.244
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr1_+_4715104
|
0.240
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr13_-_33859835
|
0.234
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_+_58906968
|
0.233
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr12_-_102874373
|
0.231
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_-_59313304
|
0.225
|
NM_001136051
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr1_-_200589856
|
0.210
|
NM_014875
|
KIF14
|
kinesin family member 14
|