Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
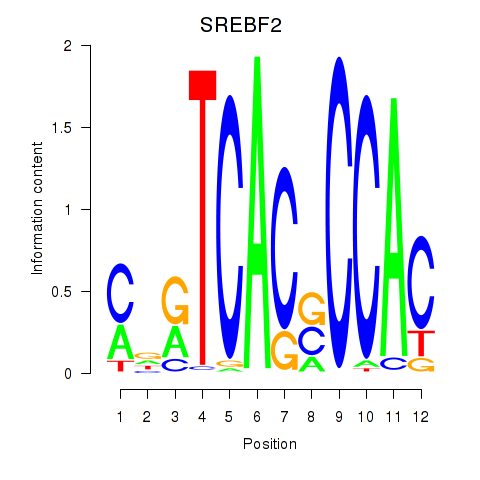
Results for SREBF2
Z-value: 1.39
Transcription factors associated with SREBF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF2
|
ENSG00000198911.7 | sterol regulatory element binding transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SREBF2 | hg19_v2_chr22_+_42229100_42229146 | 0.15 | 4.1e-01 | Click! |
Activity profile of SREBF2 motif
Sorted Z-values of SREBF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_56150184 | 4.76 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr12_-_53343602 | 4.03 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr6_+_151561085 | 3.37 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_-_53343633 | 3.26 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr9_+_96928516 | 3.16 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr12_-_91505608 | 3.01 |
ENST00000266718.4
|
LUM
|
lumican |
| chr12_-_53343560 | 2.91 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr7_-_84816122 | 2.78 |
ENST00000444867.1
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr19_+_55591743 | 2.68 |
ENST00000588359.1
ENST00000245618.5 |
EPS8L1
|
EPS8-like 1 |
| chr6_-_32152064 | 2.51 |
ENST00000375076.4
ENST00000375070.3 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr6_-_32152020 | 2.47 |
ENST00000375055.2
|
AGER
|
advanced glycosylation end product-specific receptor |
| chr15_+_47476275 | 2.43 |
ENST00000558014.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr6_-_32151999 | 2.38 |
ENST00000375069.3
ENST00000538695.1 ENST00000438221.2 ENST00000375065.5 ENST00000450110.1 ENST00000375067.3 ENST00000375056.2 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr19_-_11308190 | 2.11 |
ENST00000586659.1
ENST00000592903.1 ENST00000589359.1 ENST00000588724.1 ENST00000432929.2 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr5_-_149535421 | 2.10 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr2_-_219925189 | 1.80 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr16_+_2083265 | 1.74 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr6_+_41604747 | 1.74 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr17_-_15168624 | 1.70 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr6_-_39902160 | 1.60 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr5_+_137774706 | 1.55 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr9_-_117267717 | 1.53 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr1_+_33219592 | 1.49 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr8_-_17579726 | 1.46 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr6_+_41604620 | 1.41 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr15_+_33010175 | 1.40 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr5_+_159656437 | 1.35 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr6_+_43968306 | 1.30 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr5_-_149535190 | 1.28 |
ENST00000517488.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr3_-_160117301 | 1.19 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr16_+_22103828 | 1.15 |
ENST00000567131.1
ENST00000568328.1 ENST00000389398.5 ENST00000389397.4 |
VWA3A
|
von Willebrand factor A domain containing 3A |
| chr19_+_11200038 | 1.12 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr8_+_21882244 | 1.06 |
ENST00000289820.6
ENST00000381530.5 |
NPM2
|
nucleophosmin/nucleoplasmin 2 |
| chr6_-_52860171 | 1.06 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_-_52859968 | 0.85 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chrX_-_153236819 | 0.83 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr2_-_232571621 | 0.82 |
ENST00000595658.1
|
MGC4771
|
MGC4771 |
| chr3_-_98620500 | 0.79 |
ENST00000326840.6
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr12_-_48398104 | 0.79 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr3_-_171527560 | 0.76 |
ENST00000331659.2
|
PP13439
|
PP13439 |
| chr5_-_43313574 | 0.75 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr1_+_55505184 | 0.74 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr6_+_69942298 | 0.72 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr12_+_50344516 | 0.71 |
ENST00000199280.3
ENST00000550862.1 |
AQP2
|
aquaporin 2 (collecting duct) |
| chrX_-_153237258 | 0.68 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr16_+_66914264 | 0.66 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr4_+_185909970 | 0.64 |
ENST00000505053.1
|
RP11-386B13.4
|
RP11-386B13.4 |
| chr5_-_43313269 | 0.54 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chrX_-_103087136 | 0.51 |
ENST00000243298.2
|
RAB9B
|
RAB9B, member RAS oncogene family |
| chr19_+_36266433 | 0.50 |
ENST00000314737.5
|
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr19_+_36266417 | 0.46 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr1_+_39796810 | 0.45 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr4_+_75230853 | 0.44 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr19_-_46285646 | 0.42 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr5_-_139283982 | 0.42 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr6_-_53213780 | 0.42 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr14_-_35183886 | 0.41 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr7_-_44580861 | 0.37 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr11_+_22688150 | 0.36 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr3_-_16554403 | 0.32 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr2_+_232572361 | 0.26 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr19_-_46285736 | 0.22 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chrX_+_27826107 | 0.22 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr6_-_53213587 | 0.21 |
ENST00000542638.1
ENST00000370913.5 ENST00000541407.1 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr9_+_126131131 | 0.21 |
ENST00000373629.2
|
CRB2
|
crumbs homolog 2 (Drosophila) |
| chr6_-_32083106 | 0.21 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chrX_-_153236620 | 0.19 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr4_+_718896 | 0.19 |
ENST00000433814.1
|
PCGF3
|
polycomb group ring finger 3 |
| chr8_+_26434578 | 0.18 |
ENST00000493789.2
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr11_+_65479462 | 0.17 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr3_+_5020801 | 0.14 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr8_+_104513086 | 0.11 |
ENST00000406091.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_232573222 | 0.10 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr11_+_2923499 | 0.08 |
ENST00000449793.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr11_+_2923423 | 0.05 |
ENST00000312221.5
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr11_+_65479702 | 0.05 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr3_+_46742823 | 0.05 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr9_-_99381660 | 0.04 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr3_+_188889737 | 0.03 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr8_+_26435359 | 0.02 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr3_+_160117418 | 0.02 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr13_+_24844857 | 0.01 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr3_-_171528227 | 0.00 |
ENST00000356327.5
ENST00000342215.6 ENST00000340989.4 ENST00000351298.4 |
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:1904603 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 1.1 | 3.4 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.7 | 2.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 1.8 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.6 | 10.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.5 | 4.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 1.4 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 1.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.3 | 3.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.3 | 1.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 0.7 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 3.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 2.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.7 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.4 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 | 1.6 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 3.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.8 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 1.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 2.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 1.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 2.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.5 | 1.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.4 | 1.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.3 | 4.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 10.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 5.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 1.0 | 4.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.8 | 3.4 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.4 | 1.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 1.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 0.7 | GO:0070326 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 2.7 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 3.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 1.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 10.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 1.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 7.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 6.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 6.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 4.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |