Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
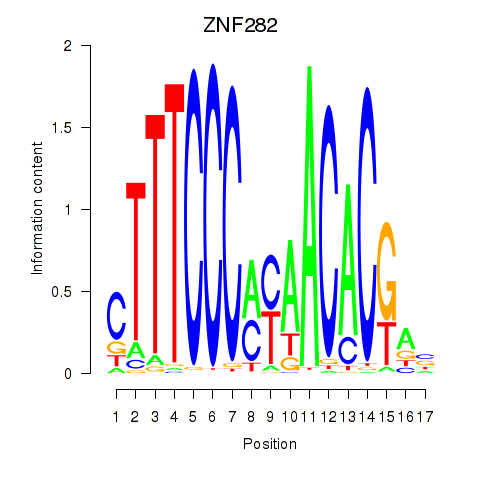
Results for ZNF282
Z-value: 1.00
Transcription factors associated with ZNF282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF282
|
ENSG00000170265.7 | zinc finger protein 282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF282 | hg19_v2_chr7_+_148892557_148892580 | -0.36 | 4.6e-02 | Click! |
Activity profile of ZNF282 motif
Sorted Z-values of ZNF282 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_36989427 | 3.38 |
ENST00000354822.5
|
NKX2-1
|
NK2 homeobox 1 |
| chr14_-_36989336 | 2.81 |
ENST00000522719.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr6_-_32151999 | 1.66 |
ENST00000375069.3
ENST00000538695.1 ENST00000438221.2 ENST00000375065.5 ENST00000450110.1 ENST00000375067.3 ENST00000375056.2 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr6_-_32152064 | 1.60 |
ENST00000375076.4
ENST00000375070.3 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr8_-_121825088 | 1.54 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr6_-_32152020 | 1.46 |
ENST00000375055.2
|
AGER
|
advanced glycosylation end product-specific receptor |
| chr19_+_18283959 | 1.41 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr12_-_113841678 | 1.21 |
ENST00000552280.1
ENST00000257549.4 |
SDS
|
serine dehydratase |
| chr17_+_7942335 | 0.88 |
ENST00000380183.4
ENST00000572022.1 ENST00000380173.2 |
ALOX15B
|
arachidonate 15-lipoxygenase, type B |
| chr1_+_2004901 | 0.83 |
ENST00000400921.2
|
PRKCZ
|
protein kinase C, zeta |
| chr1_+_2005425 | 0.81 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr5_+_167718604 | 0.80 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr12_-_127256772 | 0.77 |
ENST00000536517.1
|
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr2_-_107502456 | 0.76 |
ENST00000419159.2
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr12_+_57828521 | 0.73 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr6_+_144471643 | 0.71 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr6_+_139117251 | 0.68 |
ENST00000401414.3
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr15_+_38276910 | 0.68 |
ENST00000558081.2
|
RP11-1008C21.1
|
RP11-1008C21.1 |
| chr2_+_232260254 | 0.68 |
ENST00000287590.5
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr10_+_11865347 | 0.63 |
ENST00000277570.5
|
PROSER2
|
proline and serine-rich protein 2 |
| chr1_+_2005126 | 0.62 |
ENST00000495347.1
|
PRKCZ
|
protein kinase C, zeta |
| chr6_+_10585979 | 0.59 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr6_+_148593425 | 0.55 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr11_-_321340 | 0.52 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr1_+_226250379 | 0.52 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr17_+_7942424 | 0.50 |
ENST00000573359.1
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B |
| chr4_-_74904398 | 0.49 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr8_+_74888417 | 0.48 |
ENST00000517439.1
ENST00000312184.5 |
TMEM70
|
transmembrane protein 70 |
| chr16_+_50727479 | 0.45 |
ENST00000531674.1
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr9_+_126777676 | 0.45 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr3_+_10312604 | 0.40 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr12_-_46766577 | 0.39 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr22_-_50970919 | 0.37 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_-_79905109 | 0.36 |
ENST00000409745.2
|
MYADML2
|
myeloid-associated differentiation marker-like 2 |
| chr13_+_31309645 | 0.33 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr10_+_51572408 | 0.32 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr22_-_50970566 | 0.32 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_-_47786375 | 0.32 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr6_+_139349903 | 0.31 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr5_+_66675200 | 0.30 |
ENST00000503106.1
|
RP11-434D9.1
|
RP11-434D9.1 |
| chr1_-_24151892 | 0.30 |
ENST00000235958.4
|
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr11_+_111957497 | 0.30 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chrX_-_47004878 | 0.30 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr5_+_175299743 | 0.30 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr10_+_51572339 | 0.29 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_-_24151903 | 0.29 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr2_+_219283815 | 0.28 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr5_+_7869217 | 0.28 |
ENST00000264668.2
ENST00000514220.1 ENST00000341013.6 ENST00000440940.2 ENST00000502550.1 ENST00000506877.1 |
MTRR
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr1_+_27320176 | 0.28 |
ENST00000522111.2
|
TRNP1
|
TMF1-regulated nuclear protein 1 |
| chr11_+_100784231 | 0.28 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr22_-_50970506 | 0.28 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr2_-_188378368 | 0.28 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr8_+_124968225 | 0.28 |
ENST00000399018.1
|
FER1L6
|
fer-1-like 6 (C. elegans) |
| chr12_-_122296755 | 0.28 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr6_+_32938692 | 0.28 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr12_-_127256858 | 0.27 |
ENST00000542248.1
ENST00000540684.1 |
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr21_+_35747773 | 0.27 |
ENST00000399292.3
ENST00000399299.1 ENST00000399295.2 |
SMIM11
|
small integral membrane protein 11 |
| chr18_+_43753500 | 0.27 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr20_-_62493217 | 0.27 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr1_+_47799446 | 0.26 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr5_+_156887027 | 0.26 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr6_+_32938665 | 0.26 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr18_-_54318353 | 0.24 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr11_-_6633799 | 0.24 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr1_+_47799542 | 0.24 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr16_+_29802036 | 0.23 |
ENST00000561482.1
ENST00000160827.4 ENST00000569636.2 ENST00000400750.2 |
KIF22
|
kinesin family member 22 |
| chr5_-_175788758 | 0.23 |
ENST00000510164.1
ENST00000533553.1 ENST00000504688.1 ENST00000393725.2 ENST00000503082.1 ENST00000506983.1 |
KIAA1191
|
KIAA1191 |
| chr3_-_180707306 | 0.21 |
ENST00000479269.1
|
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr6_-_79944336 | 0.21 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr17_+_42422629 | 0.20 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chrX_-_13835147 | 0.20 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr4_+_52709229 | 0.20 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr19_+_7953384 | 0.19 |
ENST00000306708.6
|
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr3_+_45928029 | 0.19 |
ENST00000422395.1
ENST00000355983.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr17_-_73389854 | 0.18 |
ENST00000578961.1
ENST00000392564.1 ENST00000582582.1 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr1_+_35247859 | 0.17 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr14_+_71788096 | 0.17 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr6_+_10748019 | 0.17 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr11_+_82904746 | 0.17 |
ENST00000393389.3
ENST00000528722.1 |
ANKRD42
|
ankyrin repeat domain 42 |
| chr18_-_47018869 | 0.17 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr17_-_73937028 | 0.17 |
ENST00000586631.2
|
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chr10_+_114135004 | 0.17 |
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr19_-_51340469 | 0.16 |
ENST00000326856.4
|
KLK15
|
kallikrein-related peptidase 15 |
| chr10_+_95753714 | 0.16 |
ENST00000260766.3
|
PLCE1
|
phospholipase C, epsilon 1 |
| chrX_-_13835461 | 0.16 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr17_+_42422637 | 0.15 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr18_-_47017956 | 0.15 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr2_+_233415488 | 0.15 |
ENST00000454501.1
|
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr2_+_233415363 | 0.14 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr20_-_2451395 | 0.14 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr6_+_10747986 | 0.14 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr12_+_4758264 | 0.14 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr6_+_139349817 | 0.14 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr19_+_39390587 | 0.14 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_-_126480381 | 0.13 |
ENST00000368836.2
|
METTL10
|
methyltransferase like 10 |
| chr18_-_47018897 | 0.13 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr13_+_50656307 | 0.12 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr18_-_47018769 | 0.12 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr1_-_154178803 | 0.12 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr8_-_28347737 | 0.12 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr14_+_55590646 | 0.12 |
ENST00000553493.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr12_-_9102549 | 0.11 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr17_+_42422662 | 0.11 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr6_-_17706618 | 0.11 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr10_-_114206649 | 0.11 |
ENST00000369404.3
ENST00000369405.3 |
ZDHHC6
|
zinc finger, DHHC-type containing 6 |
| chr3_-_170588163 | 0.10 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr11_+_66278080 | 0.10 |
ENST00000318312.7
ENST00000526815.1 ENST00000537537.1 ENST00000525809.1 ENST00000455748.2 ENST00000393994.2 |
BBS1
|
Bardet-Biedl syndrome 1 |
| chr2_+_108602971 | 0.10 |
ENST00000409059.1
ENST00000540517.1 ENST00000264047.2 |
SLC5A7
|
solute carrier family 5 (sodium/choline cotransporter), member 7 |
| chr13_-_95248511 | 0.10 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr4_+_88343952 | 0.09 |
ENST00000440591.2
|
NUDT9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr15_-_44955842 | 0.09 |
ENST00000427534.2
ENST00000559193.1 ENST00000261866.7 ENST00000535302.2 ENST00000558319.1 |
SPG11
|
spastic paraplegia 11 (autosomal recessive) |
| chr8_-_75233563 | 0.09 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr4_+_88343717 | 0.09 |
ENST00000302174.4
ENST00000512216.1 ENST00000473942.1 |
NUDT9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr19_+_30097181 | 0.09 |
ENST00000586420.1
ENST00000221770.3 ENST00000392279.3 ENST00000590688.1 |
POP4
|
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr17_-_61996160 | 0.09 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chr19_+_44617511 | 0.08 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr2_-_9695847 | 0.08 |
ENST00000310823.3
ENST00000497134.1 |
ADAM17
|
ADAM metallopeptidase domain 17 |
| chr1_-_248738080 | 0.08 |
ENST00000328782.2
|
OR2T34
|
olfactory receptor, family 2, subfamily T, member 34 |
| chr1_+_160313062 | 0.08 |
ENST00000294785.5
ENST00000368063.1 ENST00000437169.1 |
NCSTN
|
nicastrin |
| chr3_-_170587974 | 0.08 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr18_+_54318893 | 0.08 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr19_-_58220517 | 0.08 |
ENST00000512439.2
ENST00000426889.1 |
ZNF154
|
zinc finger protein 154 |
| chr1_-_2126174 | 0.07 |
ENST00000400919.3
ENST00000420515.1 ENST00000378543.2 ENST00000400918.3 |
C1orf86
|
chromosome 1 open reading frame 86 |
| chr1_+_145507587 | 0.07 |
ENST00000330165.8
ENST00000369307.3 |
RBM8A
|
RNA binding motif protein 8A |
| chr1_-_2126192 | 0.07 |
ENST00000378546.4
|
C1orf86
|
chromosome 1 open reading frame 86 |
| chr18_+_54318566 | 0.06 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr16_+_66586461 | 0.06 |
ENST00000264001.4
ENST00000351137.4 ENST00000345436.4 ENST00000362093.4 ENST00000417030.2 ENST00000527729.1 ENST00000532838.1 |
CKLF
CKLF-CMTM1
|
chemokine-like factor CKLF-CMTM1 readthrough |
| chr2_+_242289502 | 0.06 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr18_+_54318616 | 0.06 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr1_+_150122034 | 0.06 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr12_+_93964158 | 0.05 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr2_-_68479614 | 0.05 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr19_+_39390320 | 0.04 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_16828866 | 0.04 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr17_-_27188984 | 0.04 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr5_-_7869108 | 0.04 |
ENST00000264669.5
ENST00000507572.1 ENST00000504695.1 |
FASTKD3
|
FAST kinase domains 3 |
| chr7_+_139026057 | 0.04 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr20_-_62284766 | 0.03 |
ENST00000370053.1
|
STMN3
|
stathmin-like 3 |
| chr4_+_39460659 | 0.03 |
ENST00000513731.1
|
LIAS
|
lipoic acid synthetase |
| chr10_+_114206956 | 0.01 |
ENST00000432306.1
ENST00000393077.2 |
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A |
| chr16_+_30207122 | 0.01 |
ENST00000395137.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_-_108735440 | 0.01 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr17_+_41323204 | 0.01 |
ENST00000542611.1
ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr14_+_58711539 | 0.01 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr16_+_29467780 | 0.01 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr9_+_34652164 | 0.01 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr12_+_122150646 | 0.00 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr15_-_83736091 | 0.00 |
ENST00000261721.4
|
BTBD1
|
BTB (POZ) domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF282
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1904596 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.5 | 6.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 2.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 1.4 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.5 | GO:0060585 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.5 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.5 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.7 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.3 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0046597 | response to interferon-alpha(GO:0035455) negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.8 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 1.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 4.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0005655 | ribonuclease MRP complex(GO:0000172) nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.4 | 1.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.3 | 1.4 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.3 | 6.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.6 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 1.6 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 6.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |