Project
Illumina Body Map 2
Navigation
Downloads
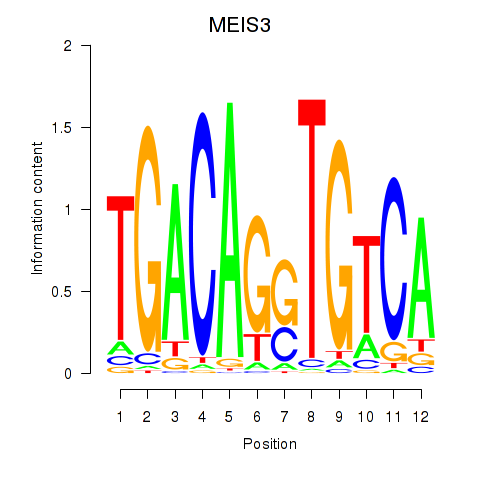
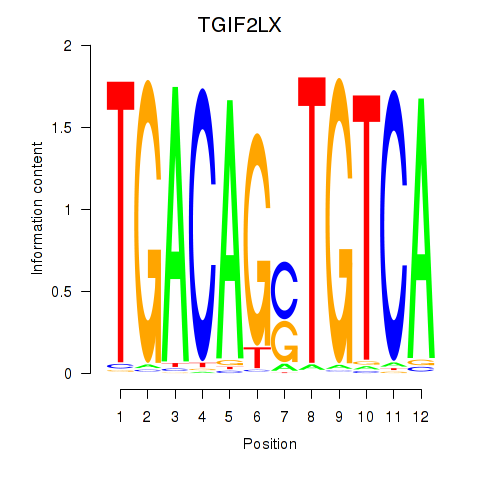
Results for MEIS3_TGIF2LX
Z-value: 1.15
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2LX | hg19_v2_chrX_+_89176881_89176955 | 0.15 | 4.0e-01 | Click! |
| MEIS3 | hg19_v2_chr19_-_47922373_47922572 | -0.13 | 4.7e-01 | Click! |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_107590383 | 5.91 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr8_+_86351056 | 4.58 |
ENST00000285381.2
|
CA3
|
carbonic anhydrase III, muscle specific |
| chr19_-_45826125 | 4.37 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr17_+_41003166 | 4.12 |
ENST00000308423.2
|
AOC3
|
amine oxidase, copper containing 3 |
| chr3_+_8775466 | 3.91 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr11_+_18287721 | 3.84 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr11_+_18287801 | 3.81 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr12_-_54978086 | 3.63 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr17_-_1395954 | 3.02 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr16_+_1290694 | 2.63 |
ENST00000338844.3
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr16_+_1290725 | 2.63 |
ENST00000461509.2
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr11_+_117947782 | 2.51 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_-_47655686 | 2.45 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr5_+_115298165 | 2.32 |
ENST00000357872.4
|
AQPEP
|
Aminopeptidase Q |
| chr1_-_201081579 | 2.26 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr11_+_1942580 | 2.18 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr16_+_1306060 | 1.99 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chr9_-_22009297 | 1.96 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr11_+_120110863 | 1.90 |
ENST00000543440.2
|
POU2F3
|
POU class 2 homeobox 3 |
| chr10_-_5060147 | 1.85 |
ENST00000604507.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chrX_+_135618258 | 1.85 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr10_-_5060201 | 1.80 |
ENST00000407674.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr9_-_22009241 | 1.78 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr2_-_42160486 | 1.78 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr11_-_2170786 | 1.76 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr5_+_115298145 | 1.69 |
ENST00000395528.2
|
AQPEP
|
Aminopeptidase Q |
| chr11_-_119187826 | 1.67 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr16_+_66410244 | 1.55 |
ENST00000562048.1
ENST00000568155.2 |
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr17_-_39781054 | 1.51 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr17_+_37782955 | 1.48 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr3_+_154801678 | 1.45 |
ENST00000462837.1
|
MME
|
membrane metallo-endopeptidase |
| chr16_+_1306093 | 1.42 |
ENST00000211076.3
|
TPSD1
|
tryptase delta 1 |
| chr8_+_67039278 | 1.42 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr11_-_76381029 | 1.41 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr17_-_39780634 | 1.40 |
ENST00000577817.2
|
KRT17
|
keratin 17 |
| chr6_-_150219232 | 1.40 |
ENST00000531073.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr10_+_99349450 | 1.40 |
ENST00000370640.3
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr9_+_34990219 | 1.35 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_-_7661623 | 1.35 |
ENST00000298441.6
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr4_-_187517928 | 1.33 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr6_+_160769300 | 1.30 |
ENST00000275300.2
|
SLC22A3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr8_+_67039131 | 1.27 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr5_-_42887494 | 1.26 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr6_+_160769399 | 1.22 |
ENST00000392145.1
|
SLC22A3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr21_-_43735446 | 1.21 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr2_+_97203082 | 1.20 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr18_+_7231123 | 1.19 |
ENST00000383467.2
|
LRRC30
|
leucine rich repeat containing 30 |
| chr1_+_46379254 | 1.19 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr5_-_44388899 | 1.16 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chr2_+_97202480 | 1.14 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr10_+_50507232 | 1.11 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr19_-_51529849 | 1.10 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr1_-_153517473 | 1.09 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr10_+_123970670 | 1.07 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_-_34122596 | 1.07 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr17_-_39780819 | 1.06 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr19_-_51530916 | 1.05 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr4_-_18023350 | 1.05 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr12_+_95611536 | 1.05 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_86934526 | 1.05 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr11_+_827553 | 1.04 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr18_+_3449330 | 1.03 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr13_+_78109884 | 1.02 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr12_-_53298841 | 1.01 |
ENST00000293308.6
|
KRT8
|
keratin 8 |
| chr19_-_51531272 | 1.01 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr16_-_2004683 | 0.99 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr12_+_95611516 | 0.98 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_51531210 | 0.97 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr13_+_78109804 | 0.96 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr9_-_86432547 | 0.96 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr17_-_19648916 | 0.94 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr2_-_218770168 | 0.91 |
ENST00000413554.1
|
TNS1
|
tensin 1 |
| chr1_-_155162658 | 0.90 |
ENST00000368389.2
ENST00000368396.4 ENST00000343256.5 ENST00000342482.4 ENST00000368398.3 ENST00000368390.3 ENST00000337604.5 ENST00000368392.3 ENST00000438413.1 ENST00000368393.3 ENST00000457295.2 ENST00000338684.5 ENST00000368395.1 |
MUC1
|
mucin 1, cell surface associated |
| chr6_+_53948328 | 0.89 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr3_+_29322437 | 0.87 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr12_+_95611569 | 0.86 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr3_-_149093499 | 0.85 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr21_+_30502806 | 0.84 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr3_-_49726486 | 0.81 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_+_86511549 | 0.81 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr6_+_53948221 | 0.80 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr18_-_52989525 | 0.79 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr7_-_141957847 | 0.77 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr14_-_67859422 | 0.76 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr1_-_57285038 | 0.75 |
ENST00000343433.6
|
C1orf168
|
chromosome 1 open reading frame 168 |
| chr22_+_21369316 | 0.74 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr11_-_65325664 | 0.73 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr17_-_42345487 | 0.71 |
ENST00000262418.6
|
SLC4A1
|
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr17_+_37783197 | 0.69 |
ENST00000582680.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr20_-_57089934 | 0.69 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr4_-_184241927 | 0.69 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr19_-_9092018 | 0.68 |
ENST00000397910.4
|
MUC16
|
mucin 16, cell surface associated |
| chr12_+_104458235 | 0.67 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr17_+_7210294 | 0.67 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_+_15500280 | 0.67 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr3_+_147795932 | 0.66 |
ENST00000490465.1
|
RP11-639B1.1
|
RP11-639B1.1 |
| chr19_-_40791211 | 0.66 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr11_-_101000445 | 0.65 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr15_+_65337708 | 0.65 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr18_+_3449821 | 0.64 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_228395755 | 0.64 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr17_+_37783170 | 0.63 |
ENST00000254079.4
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr3_+_49726932 | 0.63 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr1_+_12704566 | 0.63 |
ENST00000376221.1
|
AADACL4
|
arylacetamide deacetylase-like 4 |
| chr9_+_96026230 | 0.63 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr3_-_133748913 | 0.62 |
ENST00000310926.4
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr7_-_156803329 | 0.62 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr5_-_58882219 | 0.61 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_151662815 | 0.60 |
ENST00000359755.5
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_+_31887761 | 0.60 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chrX_-_11445856 | 0.59 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr18_+_3449695 | 0.58 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_133748758 | 0.58 |
ENST00000493729.1
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr11_+_117947724 | 0.56 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr5_+_155753745 | 0.56 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr7_+_116654958 | 0.56 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr14_+_24600484 | 0.56 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr9_-_33447584 | 0.55 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr22_+_39052632 | 0.55 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr4_+_175839506 | 0.55 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr11_-_76381781 | 0.55 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr2_-_241624705 | 0.54 |
ENST00000599492.1
|
AC011298.1
|
HCG2013738; Uncharacterized protein |
| chr4_+_175839551 | 0.53 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr11_-_842509 | 0.52 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr3_+_154801312 | 0.52 |
ENST00000497890.1
|
MME
|
membrane metallo-endopeptidase |
| chr21_+_30503282 | 0.51 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr14_-_21502944 | 0.51 |
ENST00000382951.3
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active) |
| chrX_+_23925918 | 0.51 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr9_-_140444814 | 0.50 |
ENST00000277531.4
|
PNPLA7
|
patatin-like phospholipase domain containing 7 |
| chr8_+_7345191 | 0.50 |
ENST00000335510.6
|
DEFB105B
|
defensin, beta 105B |
| chr1_-_55341551 | 0.50 |
ENST00000537443.1
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr5_-_16738451 | 0.50 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr17_+_28256874 | 0.50 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr2_+_233271546 | 0.50 |
ENST00000295453.3
|
ALPPL2
|
alkaline phosphatase, placental-like 2 |
| chr8_-_7681411 | 0.49 |
ENST00000334773.6
|
DEFB105A
|
defensin, beta 105A |
| chr17_+_73089382 | 0.49 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr17_-_76220740 | 0.47 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr1_-_153013588 | 0.46 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr1_+_82165350 | 0.46 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr3_-_52719888 | 0.46 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr19_+_6361754 | 0.45 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chrX_+_17755563 | 0.45 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr8_+_95653840 | 0.44 |
ENST00000520385.1
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr10_-_79398127 | 0.43 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_67360712 | 0.43 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr19_+_6361841 | 0.42 |
ENST00000596605.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr14_-_54418598 | 0.42 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chrX_-_117250740 | 0.42 |
ENST00000371882.1
ENST00000540167.1 ENST00000545703.1 |
KLHL13
|
kelch-like family member 13 |
| chr2_+_232457569 | 0.42 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chr6_+_112375275 | 0.42 |
ENST00000368666.2
ENST00000604763.1 ENST00000230529.5 |
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr3_-_12883026 | 0.41 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chrX_+_17755696 | 0.41 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr19_+_6361795 | 0.41 |
ENST00000596149.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr6_+_34204642 | 0.41 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr19_+_1000418 | 0.40 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr11_+_86511569 | 0.40 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr6_+_50061315 | 0.40 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr10_+_104503727 | 0.40 |
ENST00000448841.1
|
WBP1L
|
WW domain binding protein 1-like |
| chr19_-_40791302 | 0.39 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_+_145549203 | 0.39 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr2_-_175202151 | 0.39 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr19_+_48337701 | 0.38 |
ENST00000535362.1
|
TPRX2P
|
tetra-peptide repeat homeobox 2 pseudogene |
| chr6_+_50681541 | 0.37 |
ENST00000008391.3
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr22_+_44319648 | 0.37 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr19_+_5455421 | 0.37 |
ENST00000222033.4
|
ZNRF4
|
zinc and ring finger 4 |
| chr2_+_26785409 | 0.37 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr19_+_18530146 | 0.36 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chrX_+_47420516 | 0.36 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr14_+_100789669 | 0.36 |
ENST00000361529.3
ENST00000557052.1 |
SLC25A47
|
solute carrier family 25, member 47 |
| chr4_+_175839517 | 0.35 |
ENST00000502940.1
ENST00000502305.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr9_-_130667592 | 0.34 |
ENST00000447681.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr10_-_104597286 | 0.33 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr2_+_162087577 | 0.33 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_79533608 | 0.33 |
ENST00000572760.1
ENST00000573876.1 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr7_+_76139833 | 0.33 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr7_+_76139925 | 0.32 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr3_-_122512619 | 0.32 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr11_+_60524426 | 0.32 |
ENST00000528170.1
ENST00000337911.4 ENST00000405633.3 |
MS4A15
|
membrane-spanning 4-domains, subfamily A, member 15 |
| chr7_-_99332719 | 0.32 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chrY_-_2655644 | 0.31 |
ENST00000525526.2
ENST00000534739.2 ENST00000383070.1 |
SRY
|
sex determining region Y |
| chr19_+_999601 | 0.30 |
ENST00000594393.1
|
AC004528.1
|
Uncharacterized protein |
| chr2_-_3595547 | 0.29 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr1_-_32264250 | 0.29 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr11_+_47608198 | 0.29 |
ENST00000356737.2
ENST00000538490.1 |
FAM180B
|
family with sequence similarity 180, member B |
| chr17_+_48172639 | 0.28 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr11_+_45868957 | 0.28 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr4_-_135248604 | 0.27 |
ENST00000515491.1
ENST00000504728.1 ENST00000506638.1 |
RP11-400D2.2
|
RP11-400D2.2 |
| chr16_+_20817953 | 0.27 |
ENST00000568647.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr19_+_53517174 | 0.27 |
ENST00000602168.1
|
ERVV-1
|
endogenous retrovirus group V, member 1 |
| chr3_+_52719936 | 0.26 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_242089833 | 0.26 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr12_-_123215306 | 0.26 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr11_+_64073699 | 0.26 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_+_210406121 | 0.25 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr11_-_615570 | 0.25 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr19_-_10227503 | 0.25 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr19_+_18530184 | 0.25 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr2_-_65593784 | 0.25 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr11_-_18956556 | 0.25 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr7_-_144100786 | 0.25 |
ENST00000223140.5
|
NOBOX
|
NOBOX oogenesis homeobox |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.4 | 4.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.6 | 3.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 3.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 3.6 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.2 | GO:0060661 | proximal/distal axis specification(GO:0009946) bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) secretion by lung epithelial cell involved in lung growth(GO:0061033) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.3 | 4.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 2.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 2.0 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.2 | 1.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 0.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 2.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 4.0 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 7.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.2 | 1.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.6 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.7 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 2.5 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 2.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.6 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 3.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.4 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.5 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.9 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.4 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.5 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 4.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.3 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.6 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.4 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 2.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 5.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.4 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.3 | GO:2000018 | regulation of male gonad development(GO:2000018) positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.0 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.4 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 1.3 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.2 | 7.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 2.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 5.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 5.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 4.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) filopodium tip(GO:0032433) |
| 0.0 | 4.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 2.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.6 | 2.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 2.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.5 | 3.6 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.5 | 4.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 4.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 3.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 3.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 4.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.5 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.2 | 3.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 3.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 8.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 1.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 2.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 2.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 1.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 20.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 7.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 5.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 2.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 6.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 4.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 7.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 5.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 6.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 3.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |