Project
Illumina Body Map 2
Navigation
Downloads
Results for SIX6
Z-value: 1.21
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX6 | hg19_v2_chr14_+_60975644_60975673 | -0.18 | 3.2e-01 | Click! |
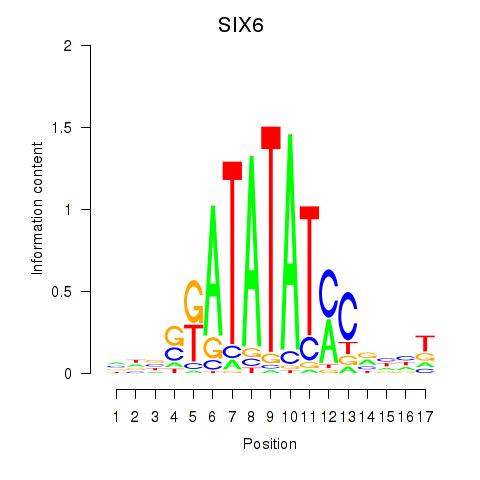
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88427568 | 11.73 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr1_-_161193349 | 6.30 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr9_-_100707116 | 6.12 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chr4_-_155511887 | 5.92 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr19_-_16008880 | 5.10 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr3_+_151531859 | 4.49 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr1_+_159557607 | 3.40 |
ENST00000255040.2
|
APCS
|
amyloid P component, serum |
| chr6_+_37012607 | 3.19 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr2_+_34935472 | 3.19 |
ENST00000604250.1
|
AC073218.2
|
AC073218.2 |
| chr3_+_151531810 | 3.19 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr8_+_67405438 | 3.16 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr13_-_46679185 | 3.15 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_+_86934526 | 3.06 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr3_+_186742464 | 3.04 |
ENST00000416235.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_63063152 | 3.03 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr15_+_58430368 | 2.95 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr13_-_46679144 | 2.87 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr16_+_72090053 | 2.84 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr15_+_58430567 | 2.80 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr1_-_159684371 | 2.76 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr7_+_117251671 | 2.76 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr12_-_14849470 | 2.66 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr6_+_25727046 | 2.46 |
ENST00000274764.2
|
HIST1H2BA
|
histone cluster 1, H2ba |
| chr1_+_21880560 | 2.45 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr4_-_71532668 | 1.75 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr6_-_25726781 | 1.70 |
ENST00000297012.3
|
HIST1H2AA
|
histone cluster 1, H2aa |
| chr4_-_71532601 | 1.69 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr7_-_92855762 | 1.58 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr22_+_18721427 | 1.57 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr8_+_101170134 | 1.56 |
ENST00000520643.1
|
SPAG1
|
sperm associated antigen 1 |
| chr16_-_20364030 | 1.49 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr15_-_80263506 | 1.47 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr15_+_69857515 | 1.46 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_-_184241927 | 1.42 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr10_-_14614311 | 1.35 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr22_+_18043133 | 1.33 |
ENST00000327451.6
ENST00000399813.1 |
SLC25A18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr8_+_101170257 | 1.33 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr19_-_14628645 | 1.29 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_-_108145499 | 1.29 |
ENST00000369020.3
ENST00000369022.2 |
SCML4
|
sex comb on midleg-like 4 (Drosophila) |
| chr5_-_66492562 | 1.28 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr4_+_74347400 | 1.27 |
ENST00000226355.3
|
AFM
|
afamin |
| chr16_-_20364122 | 1.26 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chrX_+_12809463 | 1.23 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr18_+_55018044 | 1.22 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr11_+_126262027 | 1.22 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr14_+_20937538 | 1.21 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr9_-_23821842 | 1.13 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_+_9203855 | 1.10 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr2_+_21444025 | 1.08 |
ENST00000435237.1
ENST00000457901.1 |
AC067959.1
|
AC067959.1 |
| chr2_-_239140011 | 1.07 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr14_+_83108955 | 0.99 |
ENST00000555798.1
ENST00000553760.1 ENST00000555150.1 ENST00000556970.1 |
RP11-406A9.2
|
RP11-406A9.2 |
| chr13_-_62001982 | 0.98 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr2_+_210636697 | 0.94 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr12_+_10705960 | 0.92 |
ENST00000544591.1
|
RP11-291B21.2
|
RP11-291B21.2 |
| chr11_-_45307817 | 0.91 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chrY_-_16098393 | 0.91 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chr18_+_50278430 | 0.86 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr12_-_87232771 | 0.86 |
ENST00000550014.1
|
RP11-202H2.1
|
RP11-202H2.1 |
| chr16_-_52119019 | 0.85 |
ENST00000561513.1
ENST00000565742.1 |
LINC00919
|
long intergenic non-protein coding RNA 919 |
| chr3_+_125694347 | 0.84 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr14_+_21152259 | 0.80 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chrY_+_16168097 | 0.79 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr8_+_24241789 | 0.76 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chrX_+_142113704 | 0.73 |
ENST00000446864.1
ENST00000370504.3 |
SPANXN4
|
SPANX family, member N4 |
| chr19_-_53794875 | 0.72 |
ENST00000426466.1
|
BIRC8
|
baculoviral IAP repeat containing 8 |
| chr1_+_40942887 | 0.72 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr6_-_8102279 | 0.70 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr16_+_19467772 | 0.70 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chrX_+_8433376 | 0.67 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr11_+_59807748 | 0.64 |
ENST00000278855.2
ENST00000532905.1 |
PLAC1L
|
oocyte secreted protein 2 |
| chr8_+_24241969 | 0.64 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr5_-_83016632 | 0.63 |
ENST00000515590.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_134326009 | 0.62 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr22_-_17302589 | 0.61 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr17_-_42327236 | 0.59 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr12_-_121477039 | 0.58 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr20_-_4990931 | 0.58 |
ENST00000379333.1
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chrX_-_77225135 | 0.57 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr7_-_141957847 | 0.56 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr4_-_42154895 | 0.55 |
ENST00000502486.1
ENST00000504360.1 |
BEND4
|
BEN domain containing 4 |
| chr8_+_32579341 | 0.53 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr6_+_80816342 | 0.51 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chrX_-_18238999 | 0.50 |
ENST00000380033.4
ENST00000380030.3 |
BEND2
|
BEN domain containing 2 |
| chr12_+_65277536 | 0.50 |
ENST00000540024.1
|
RP11-766N7.3
|
RP11-766N7.3 |
| chr4_-_71532339 | 0.49 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr19_+_46000479 | 0.48 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr8_-_82598067 | 0.48 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr1_+_112016414 | 0.48 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr7_+_6121296 | 0.48 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr2_-_77749387 | 0.47 |
ENST00000409884.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_+_12718497 | 0.46 |
ENST00000379348.2
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr3_-_107596910 | 0.45 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr5_+_171621176 | 0.44 |
ENST00000398186.4
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr12_+_19282643 | 0.43 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_-_87232644 | 0.42 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_53308398 | 0.41 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr3_-_146323019 | 0.40 |
ENST00000492200.1
ENST00000482567.1 |
PLSCR5
|
phospholipid scramblase family, member 5 |
| chr8_-_110660975 | 0.40 |
ENST00000528045.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr19_-_44031375 | 0.40 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr9_-_99382065 | 0.39 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr2_-_197457335 | 0.39 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr12_+_103545593 | 0.39 |
ENST00000547418.1
|
RP11-552I14.1
|
Uncharacterized protein |
| chr1_-_161015752 | 0.38 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr16_-_87350970 | 0.38 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr1_-_182369751 | 0.37 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr5_+_140201183 | 0.37 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr4_-_13546632 | 0.36 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr11_-_102496063 | 0.36 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr17_+_61271355 | 0.36 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_124789146 | 0.36 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr15_+_63414760 | 0.34 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr8_+_67104323 | 0.34 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr6_-_6711235 | 0.33 |
ENST00000432823.2
|
RP1-80N2.2
|
RP1-80N2.2 |
| chr6_+_47749718 | 0.33 |
ENST00000489301.2
ENST00000371211.2 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr6_-_26189304 | 0.33 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr12_-_122018346 | 0.31 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_-_139051839 | 0.31 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr14_+_21525981 | 0.31 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr22_-_19137796 | 0.29 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr4_+_57396766 | 0.29 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr2_-_77749336 | 0.29 |
ENST00000409282.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr12_-_498620 | 0.28 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr12_-_121476959 | 0.27 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr1_-_13673511 | 0.27 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr17_+_7788104 | 0.27 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr4_+_189321881 | 0.26 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr1_-_67142710 | 0.25 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr6_+_167562968 | 0.25 |
ENST00000486697.2
|
TCP10L2
|
t-complex 10-like 2 |
| chrY_-_24038660 | 0.25 |
ENST00000382677.3
|
RBMY1D
|
RNA binding motif protein, Y-linked, family 1, member D |
| chr5_+_115420688 | 0.24 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr5_-_78281775 | 0.24 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr6_+_127898312 | 0.24 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr13_+_34392185 | 0.23 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chrY_+_23698778 | 0.21 |
ENST00000303902.5
|
RBMY1A1
|
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr2_-_220197351 | 0.21 |
ENST00000392083.1
|
RESP18
|
regulated endocrine-specific protein 18 |
| chr1_-_10003372 | 0.21 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr21_+_22519416 | 0.20 |
ENST00000535285.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr17_-_5342380 | 0.19 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr5_+_161277603 | 0.19 |
ENST00000519621.1
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_+_156119466 | 0.18 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_+_259218 | 0.18 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr10_-_61720640 | 0.18 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr8_-_128231299 | 0.18 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr22_-_17702729 | 0.17 |
ENST00000449907.2
ENST00000441548.1 ENST00000399839.1 |
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr4_-_123377880 | 0.17 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr6_+_146348782 | 0.16 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr11_+_33902189 | 0.14 |
ENST00000330381.2
|
AC132216.1
|
HCG1785179; PRO1787; Uncharacterized protein |
| chr8_+_107593198 | 0.14 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr11_+_6149840 | 0.14 |
ENST00000316517.2
|
OR56B3P
|
olfactory receptor, family 56, subfamily B, member 3 pseudogene |
| chr3_+_40141502 | 0.13 |
ENST00000539167.1
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr1_-_36235559 | 0.13 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr2_-_58468437 | 0.11 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr1_-_161015663 | 0.10 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr12_+_122018697 | 0.10 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr8_+_32579271 | 0.09 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr6_+_146348810 | 0.09 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr2_-_77749474 | 0.09 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr1_+_234509186 | 0.09 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr2_+_181988560 | 0.08 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr12_-_122018859 | 0.07 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr17_+_60501228 | 0.06 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr1_+_179263308 | 0.06 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr2_+_177134201 | 0.05 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr2_+_177134134 | 0.05 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr19_+_31658405 | 0.05 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr2_-_89266286 | 0.04 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr13_-_20080080 | 0.03 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr4_+_74301880 | 0.02 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr14_+_22948510 | 0.02 |
ENST00000390483.1
|
TRAJ56
|
T cell receptor alpha joining 56 |
| chr9_-_21142144 | 0.02 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr5_+_120649672 | 0.01 |
ENST00000609647.1
|
CTC-546K23.1
|
CTC-546K23.1 |
| chr20_+_54987305 | 0.00 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 1.7 | 5.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 1.5 | 6.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.0 | 5.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.9 | 2.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.8 | 2.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.7 | 11.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.7 | 2.8 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.6 | 7.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.5 | 2.7 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 3.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.4 | 5.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 1.2 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.4 | 3.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 2.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 1.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 2.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.9 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 0.6 | GO:0015993 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 1.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 0.8 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 2.8 | GO:0008228 | opsonization(GO:0008228) positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 3.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.2 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.3 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 | 0.5 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 2.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 2.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.6 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 1.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 5.6 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 3.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.7 | 11.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 2.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.4 | 5.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 6.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 1.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 2.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 3.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 2.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 3.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 2.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.7 | 11.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.6 | 6.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.1 | 5.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.7 | 2.8 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.6 | 6.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 2.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 2.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 3.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 7.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 0.5 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 6.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 3.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.5 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 3.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 5.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 10.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |