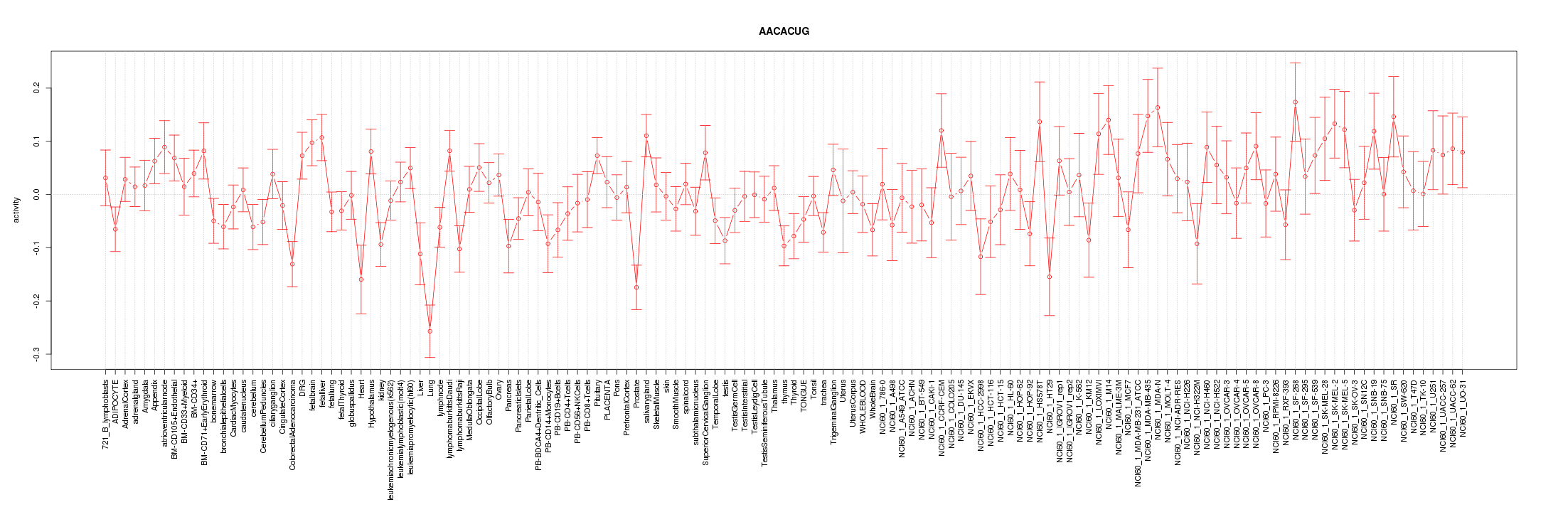
|
chr3_-_195808958
|
14.147
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_195808997
|
13.921
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_-_150207025
|
11.058
|
NM_001136479
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr1_-_150208462
|
10.056
|
NM_001136478
NM_030920
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr6_-_18264638
|
9.923
|
NM_001134709
NM_003472
|
DEK
|
DEK oncogene
|
|
chr6_+_11538486
|
9.545
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr8_+_26149004
|
8.088
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr17_-_8534008
|
7.822
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr8_-_12973752
|
7.801
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_+_26150731
|
7.005
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr8_+_20054703
|
6.546
|
NM_001693
|
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2
|
|
chr5_+_112312392
|
6.239
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr13_+_98605928
|
6.199
|
NM_002271
|
IPO5
|
importin 5
|
|
chr12_-_54653369
|
6.182
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr12_-_54653317
|
6.157
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr6_+_163835668
|
6.061
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr5_+_145826866
|
5.839
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr2_+_178257371
|
5.702
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr1_+_40723730
|
5.688
|
NM_005857
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr2_+_160568973
|
5.574
|
NM_022826
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr5_+_179125796
|
5.544
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr6_+_64281910
|
5.523
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr4_+_146019155
|
5.494
|
NM_001040876
NM_002940
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr5_+_72143929
|
5.421
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr11_+_86748870
|
5.273
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr8_-_12990731
|
5.255
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_-_149375584
|
5.251
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr18_-_13726582
|
5.211
|
NM_001098801
NM_152352
|
FAM210A
|
family with sequence similarity 210, member A
|
|
chr6_-_52441815
|
5.152
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_+_84609941
|
5.107
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_-_29151724
|
5.102
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr15_+_77713229
|
5.016
|
NM_018200
|
HMG20A
|
high mobility group 20A
|
|
chr1_-_6445737
|
4.974
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr6_+_20402040
|
4.866
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr7_+_128379345
|
4.693
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr5_+_149340287
|
4.553
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr1_-_6420719
|
4.537
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr12_-_46766563
|
4.528
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr5_+_72112417
|
4.420
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr3_+_38206968
|
4.417
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr1_+_84630014
|
4.373
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_-_6418966
|
4.343
|
NM_181866
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr14_+_103801160
|
4.310
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr12_+_12938540
|
4.306
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr12_-_109125289
|
4.262
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr1_+_52082545
|
4.227
|
NM_024586
NM_148906
NM_148908
NM_148909
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr1_+_84647342
|
4.211
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_+_12878718
|
4.206
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr19_+_19322763
|
4.195
|
NM_004386
|
NCAN
|
neurocan
|
|
chr3_+_151985828
|
4.161
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr1_+_93811476
|
4.140
|
NM_001938
|
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2)
|
|
chr6_-_10419786
|
4.126
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr4_+_1902352
|
4.118
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr11_-_87908551
|
4.068
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr13_-_50367027
|
4.050
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr17_+_49230896
|
4.040
|
NM_000269
NM_198175
NM_001018136
|
NME1
NME1-NME2
|
non-metastatic cells 1, protein (NM23A) expressed in
NME1-NME2 readthrough
|
|
chr16_+_53088791
|
3.972
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr9_-_6015606
|
3.952
|
NM_001243202
NM_001243203
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr3_-_149421059
|
3.925
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr1_+_84630575
|
3.924
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr15_+_57210823
|
3.892
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr1_-_6453773
|
3.821
|
NM_007274
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr1_+_52195458
|
3.805
|
NM_148904
NM_148905
NM_148907
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr3_+_152017193
|
3.781
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr3_-_33700932
|
3.689
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_142720371
|
3.653
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr7_+_12726910
|
3.640
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr17_+_11924116
|
3.621
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr19_-_57336373
|
3.600
|
NM_001146186
|
PEG3
|
paternally expressed 3
|
|
chr3_+_158991035
|
3.591
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_-_20911560
|
3.578
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr2_-_37899227
|
3.498
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_-_46178559
|
3.448
|
NM_006807
|
CBX1
|
chromobox homolog 1
|
|
chr3_-_113465101
|
3.417
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr1_-_35658742
|
3.390
|
NM_005066
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr11_-_33744272
|
3.360
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr3_-_125314367
|
3.332
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr1_-_225616556
|
3.329
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr13_+_42846255
|
3.323
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr7_+_107221203
|
3.308
|
NM_001008405
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chr4_-_83719896
|
3.307
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr12_+_75784878
|
3.255
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr3_+_113465860
|
3.233
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr11_-_33757872
|
3.229
|
NM_000611
NM_203329
NM_203330
NM_203331
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr11_-_33744497
|
3.160
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr3_+_61547179
|
3.092
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr1_-_225840660
|
3.086
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr20_-_524335
|
3.085
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr2_+_113403433
|
3.059
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr2_+_102314162
|
3.049
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr15_+_57511616
|
3.035
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_-_42801529
|
2.984
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chr14_-_57735614
|
2.978
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr3_+_159557649
|
2.970
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr15_-_52821004
|
2.920
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr6_+_32121775
|
2.918
|
NM_001204103
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 readthrough
palmitoyl-protein thioesterase 2
|
|
chr3_-_185542807
|
2.914
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr4_-_71705188
|
2.889
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr3_+_160118346
|
2.867
|
NM_005496
|
SMC4
|
structural maintenance of chromosomes 4
|
|
chr7_+_107220421
|
2.850
|
NM_018844
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chr6_+_32121228
|
2.822
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr11_+_101980736
|
2.815
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_63953419
|
2.808
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr11_-_63536112
|
2.800
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr4_-_71705613
|
2.778
|
NM_002092
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr1_-_225615787
|
2.778
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr3_-_141719188
|
2.734
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chrX_+_49832214
|
2.697
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr20_+_16710608
|
2.689
|
NM_003092
NM_198220
|
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B
|
|
chr4_-_129209983
|
2.668
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr5_+_138609440
|
2.657
|
NM_001194954
NM_199189
|
MATR3
|
matrin 3
|
|
chr1_-_6614584
|
2.650
|
NM_024654
|
NOL9
|
nucleolar protein 9
|
|
chr9_+_106856531
|
2.641
|
NM_001042551
NM_006444
|
SMC2
|
structural maintenance of chromosomes 2
|
|
chr1_-_68299047
|
2.639
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr9_+_106856810
|
2.612
|
NM_001042550
|
SMC2
|
structural maintenance of chromosomes 2
|
|
chr19_+_57752051
|
2.607
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr8_-_95907423
|
2.594
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr5_-_43313581
|
2.592
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr6_-_10415438
|
2.581
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_-_37544165
|
2.572
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr17_-_46178761
|
2.523
|
NM_001127228
|
CBX1
|
chromobox homolog 1
|
|
chr8_+_71581599
|
2.516
|
NM_001011720
|
XKR9
|
XK, Kell blood group complex subunit-related family, member 9
|
|
chr12_-_76953561
|
2.503
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr8_+_55047780
|
2.477
|
NM_014175
|
MRPL15
|
mitochondrial ribosomal protein L15
|
|
chr6_+_107811272
|
2.452
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_-_133561805
|
2.403
|
NM_002715
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr4_+_57333746
|
2.400
|
NM_006947
|
SRP72
|
signal recognition particle 72kDa
|
|
chr1_-_183604885
|
2.392
|
NM_005717
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr13_-_31040035
|
2.378
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr2_-_198364623
|
2.368
|
NM_002156
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr6_+_17600504
|
2.366
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr5_+_138629419
|
2.349
|
NM_001194955
NM_001194956
|
MATR3
|
matrin 3
|
|
chr22_+_35653444
|
2.331
|
NM_001003681
|
HMGXB4
|
HMG box domain containing 4
|
|
chr20_+_60718473
|
2.329
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr2_+_187464931
|
2.324
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr2_+_223725675
|
2.283
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr15_-_25650647
|
2.278
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr16_-_71748697
|
2.277
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr9_+_112887780
|
2.275
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_+_29117503
|
2.268
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chr8_-_53626991
|
2.265
|
NM_001083617
NM_014781
|
RB1CC1
|
RB1-inducible coiled-coil 1
|
|
chr7_+_106685007
|
2.262
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr2_+_187454724
|
2.243
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr18_+_34409058
|
2.237
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr18_+_12947972
|
2.236
|
NM_001013437
NM_031216
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr20_+_5107393
|
2.221
|
NM_003818
|
CDS2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2
|
|
chr15_+_44580870
|
2.208
|
NM_138423
NM_177974
|
CASC4
|
cancer susceptibility candidate 4
|
|
chr3_-_98241824
|
2.207
|
NM_001040181
NM_001040182
NM_001040183
NM_001040199
NM_001040200
NM_019895
|
CLDND1
|
claudin domain containing 1
|
|
chr4_+_41614911
|
2.201
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_+_112810877
|
2.183
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_-_198364997
|
2.179
|
NM_199440
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr3_-_33759684
|
2.172
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_+_138629330
|
2.137
|
NM_018834
|
MATR3
|
matrin 3
|
|
chr8_-_67525426
|
2.115
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr5_+_110074740
|
2.021
|
NM_138773
|
SLC25A46
|
solute carrier family 25, member 46
|
|
chr7_+_129710336
|
2.016
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr3_-_55521330
|
2.015
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr17_+_1733263
|
2.014
|
NM_002945
|
RPA1
|
replication protein A1, 70kDa
|
|
chr10_+_31610063
|
2.004
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr1_-_11120081
|
2.002
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr2_-_161350304
|
2.001
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr11_-_107729913
|
1.991
|
NM_017515
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr9_+_112542496
|
1.981
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr3_+_133292404
|
1.978
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr3_+_133293256
|
1.969
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr1_-_205719340
|
1.968
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr1_+_116184573
|
1.968
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_+_10262853
|
1.951
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr12_-_76478737
|
1.940
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr7_-_25164902
|
1.927
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr6_-_7313352
|
1.925
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr4_+_41362750
|
1.920
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_+_11862955
|
1.911
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr11_-_110167191
|
1.898
|
NM_002906
|
RDX
|
radixin
|
|
chrX_-_119445306
|
1.884
|
NM_001104544
NM_001104545
NM_017938
|
FAM70A
|
family with sequence similarity 70, member A
|
|
chr4_+_39699663
|
1.882
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr7_-_137686814
|
1.881
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr5_+_169010637
|
1.880
|
NM_017785
|
CCDC99
|
coiled-coil domain containing 99
|
|
chr11_-_27494232
|
1.880
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr2_+_10262694
|
1.876
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr2_+_208394615
|
1.875
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr1_+_20208837
|
1.865
|
NM_015207
|
OTUD3
|
OTU domain containing 3
|
|
chr4_+_184020343
|
1.863
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr1_+_148560846
|
1.853
|
NM_173638
|
NBPF15
|
neuroblastoma breakpoint family, member 15
|
|
chr3_+_158787040
|
1.849
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr10_+_93558139
|
1.848
|
NM_025235
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr7_+_12726413
|
1.845
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr2_+_65216482
|
1.840
|
NM_003038
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr12_+_70133179
|
1.812
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr11_+_58910173
|
1.811
|
NM_001142519
NM_001142520
NM_001142521
|
FAM111A
|
family with sequence similarity 111, member A
|
|
chr15_-_91537724
|
1.807
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr6_-_10412606
|
1.806
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr10_+_31608097
|
1.804
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr3_+_63884074
|
1.802
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr2_+_65215577
|
1.793
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr9_-_123605185
|
1.788
|
NM_005047
|
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5
|
|
chr18_-_6414900
|
1.784
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chrX_-_67653169
|
1.773
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr3_-_185655833
|
1.771
|
NM_001243879
NM_004593
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr7_+_134464161
|
1.770
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|