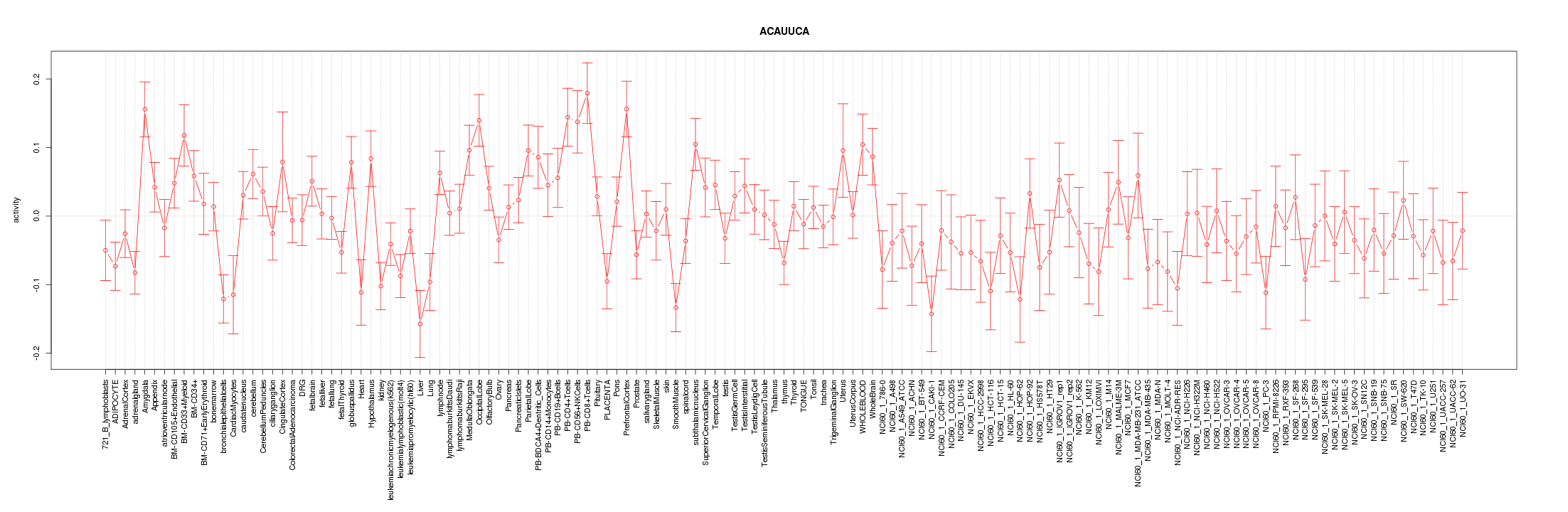
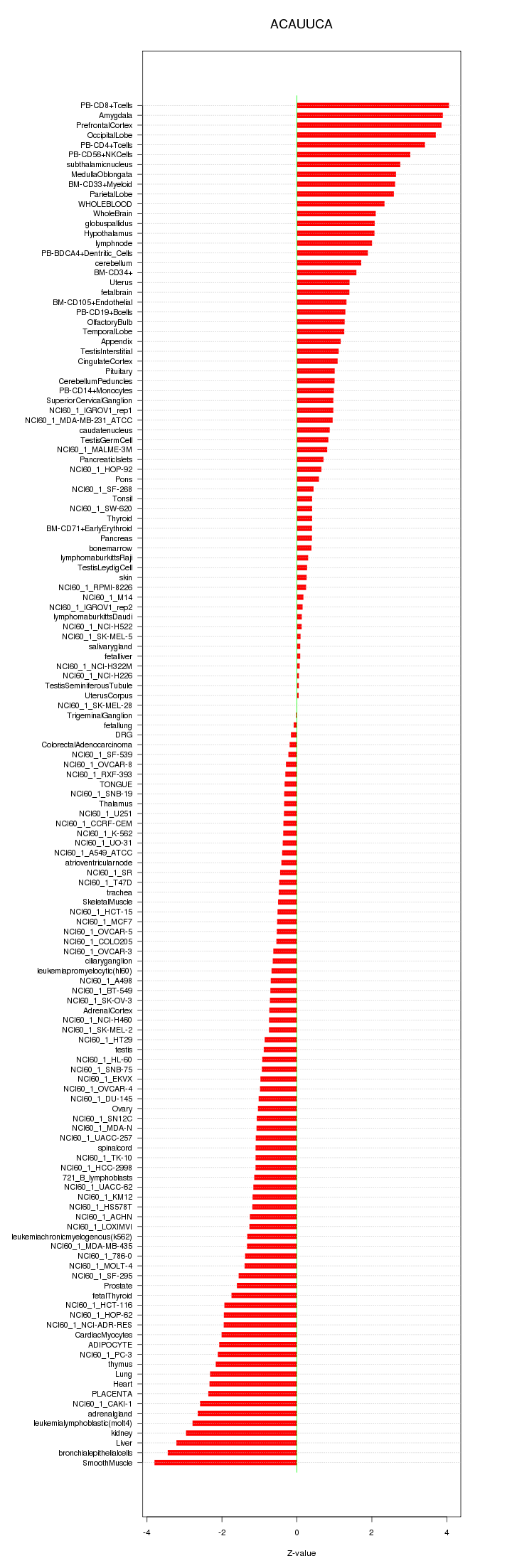
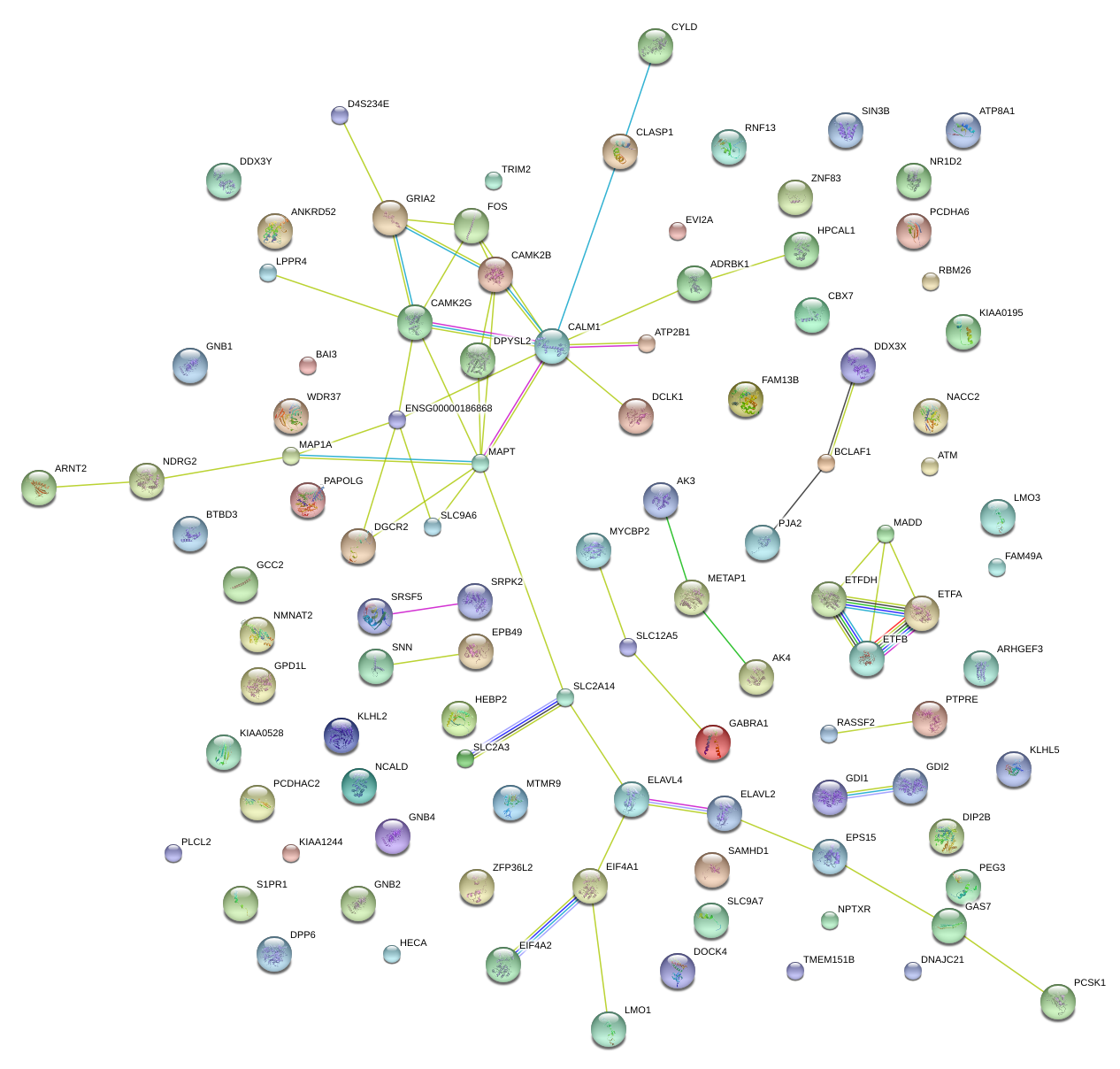
|
chr20_+_44650260
|
18.448
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr4_+_158141735
|
18.071
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr20_+_44657812
|
17.641
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr22_-_39548448
|
17.532
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr20_-_4795768
|
14.221
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr20_-_4804067
|
13.939
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr3_-_57113292
|
13.304
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_-_103136486
|
13.162
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr10_+_129845806
|
12.932
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr7_+_153584383
|
12.931
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr15_+_80696569
|
12.879
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr10_+_129705299
|
12.752
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr8_-_103137134
|
12.750
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr14_+_90863302
|
12.512
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr22_-_39240016
|
12.388
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_+_50513685
|
12.261
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_-_108745569
|
12.238
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr14_+_75745480
|
11.949
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr16_+_11762235
|
11.827
|
NM_003498
|
SNN
|
stannin
|
|
chr9_-_4726226
|
11.739
|
NM_001199856
|
AK3
|
adenylate kinase 3
|
|
chr3_-_56835829
|
11.536
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr12_-_16759531
|
11.475
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_26435339
|
11.422
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr9_-_4741798
|
11.399
|
NM_001199855
|
AK3
|
adenylate kinase 3
|
|
chr9_-_4741267
|
11.208
|
NM_001199852
NM_016282
|
AK3
|
adenylate kinase 3
|
|
chr12_-_16760935
|
11.087
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_-_4741085
|
10.869
|
NM_001199854
|
AK3
|
adenylate kinase 3
|
|
chr8_-_102803438
|
10.746
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr7_+_153749740
|
10.717
|
NM_130797
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr3_-_56809594
|
10.703
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_+_26371461
|
10.533
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_+_135067573
|
10.408
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr4_+_154125589
|
10.311
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_+_67033889
|
10.299
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr3_+_23987514
|
10.275
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_+_109065576
|
10.221
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr1_-_183387501
|
10.178
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr12_-_22697439
|
10.106
|
NM_014802
|
KIAA0528
|
KIAA0528
|
|
chr9_-_4742042
|
10.072
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr17_+_43971655
|
10.053
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr20_+_11871364
|
9.600
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr7_-_111846384
|
9.536
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr12_-_56652118
|
9.488
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr1_+_50574589
|
9.245
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr6_+_139456240
|
8.987
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr10_-_75634219
|
8.758
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr4_+_154074269
|
8.549
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr9_-_138987096
|
8.510
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr1_-_183274008
|
8.508
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr4_+_39046198
|
8.426
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr17_-_29648631
|
8.413
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr6_-_99873129
|
8.269
|
NM_015491
NM_032870
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chrX_+_153665258
|
8.063
|
NM_001493
|
GDI1
|
GDP dissociation inhibitor 1
|
|
chr7_+_154002194
|
7.982
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr11_+_47291142
|
7.980
|
NM_001135944
NM_003682
NM_130471
NM_130472
NM_130473
NM_130474
|
MADD
|
MAP-kinase activating death domain
|
|
chr5_+_161275541
|
7.976
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_50569581
|
7.832
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_+_161274196
|
7.753
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr12_-_8088629
|
7.674
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr10_+_1102741
|
7.647
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr8_+_21911062
|
7.626
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr6_+_44238479
|
7.571
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr19_+_16940172
|
7.552
|
NM_015260
|
SIN3B
|
SIN3 transcription regulator homolog B (yeast)
|
|
chr12_-_90049818
|
7.526
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr5_+_161274684
|
7.432
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_-_95767862
|
7.368
|
NM_001177875
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr4_-_42658883
|
7.306
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr3_+_149530436
|
7.247
|
NM_007282
NM_183381
|
RNF13
|
ring finger protein 13
|
|
chr13_-_36705513
|
7.182
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr8_+_21915371
|
7.007
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr13_-_36429798
|
6.719
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr5_+_161274939
|
6.697
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr11_+_47291659
|
6.696
|
NM_001135943
NM_130475
NM_130476
|
MADD
|
MAP-kinase activating death domain
|
|
chr5_+_140345746
|
6.423
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr19_-_57336373
|
6.400
|
NM_001146186
|
PEG3
|
paternally expressed 3
|
|
chr17_-_10101854
|
6.391
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr8_+_11141983
|
6.368
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr7_-_105029321
|
6.352
|
NM_182692
|
SRPK2
|
SRSF protein kinase 2
|
|
chr3_+_32148000
|
6.245
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr12_+_50898744
|
6.186
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr2_-_43453737
|
6.080
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr8_+_21916685
|
6.048
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr1_+_50571963
|
6.040
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_9929567
|
6.032
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr4_+_4387982
|
6.012
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr13_-_77901176
|
5.994
|
NM_015057
|
MYCBP2
|
MYC binding protein 2
|
|
chr5_+_161277702
|
5.983
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161277391
|
5.897
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr22_-_19109912
|
5.882
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr16_+_50776012
|
5.846
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr14_-_21493910
|
5.783
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr11_+_47290893
|
5.730
|
NM_130470
|
MADD
|
MAP-kinase activating death domain
|
|
chr8_+_21912327
|
5.723
|
NM_001114136
NM_001114139
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr19_-_53141592
|
5.700
|
NM_001105549
NM_001105550
NM_001105551
NM_001105552
NM_001105553
NM_001105554
NM_018300
|
ZNF83
|
zinc finger protein 83
|
|
chrY_+_15016698
|
5.696
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr16_+_50775960
|
5.695
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr20_-_35580186
|
5.633
|
NM_015474
|
SAMHD1
|
SAM domain and HD domain 1
|
|
chr3_+_186501360
|
5.633
|
NM_001967
|
EIF4A2
|
eukaryotic translation initiation factor 4A2
|
|
chr11_+_108093558
|
5.612
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr17_-_9862765
|
5.579
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr2_-_122406994
|
5.574
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr2_+_60983354
|
5.569
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr5_+_34929679
|
5.557
|
NM_001012339
NM_194283
|
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21
|
|
chr4_+_166128769
|
5.546
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr1_-_51984994
|
5.539
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr3_+_16926451
|
5.490
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr2_-_16847070
|
5.466
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr5_+_140254930
|
5.443
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr13_-_79979913
|
5.426
|
NM_022118
|
RBM26
|
RNA binding motif protein 26
|
|
chrY_+_15016018
|
5.356
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr14_+_70233812
|
5.354
|
NM_001039465
NM_006925
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr5_-_137368778
|
5.345
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr17_+_73452663
|
5.331
|
NM_014738
|
KIAA0195
|
KIAA0195
|
|
chr6_-_136610932
|
5.284
|
NM_001077440
NM_001077441
NM_014739
|
BCLAF1
|
BCL2-associated transcription factor 1
|
|
chr1_+_101702304
|
5.219
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr5_+_140247767
|
5.200
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr6_+_69344939
|
5.170
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr1_+_99729847
|
5.041
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr3_-_179169330
|
5.022
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr15_+_43809805
|
4.968
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr6_+_138483024
|
4.915
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr2_+_242641310
|
4.884
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr5_+_140186658
|
4.834
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_-_95748688
|
4.821
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr9_+_91933384
|
4.806
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr3_-_50540853
|
4.787
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr19_-_53193736
|
4.776
|
NM_001242531
NM_001242538
|
ZNF83
|
zinc finger protein 83
|
|
chr14_-_21493122
|
4.766
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr2_+_68592317
|
4.755
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr4_+_166131170
|
4.697
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr3_-_195163575
|
4.659
|
NM_012287
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2
|
|
chr10_+_112679277
|
4.526
|
NM_007373
|
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans)
|
|
chr5_+_140213968
|
4.496
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr3_+_23986735
|
4.491
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_20081523
|
4.424
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr18_-_5543967
|
4.378
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr3_+_159557649
|
4.336
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr5_+_161275897
|
4.335
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_65017993
|
4.249
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr5_+_140220811
|
4.248
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr16_+_50186809
|
4.236
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr5_-_95768660
|
4.213
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr9_-_91793681
|
4.194
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr3_+_45730677
|
4.191
|
NM_014016
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr9_-_123476578
|
4.173
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr17_-_9940004
|
4.166
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr14_+_105780751
|
4.133
|
NM_001100913
NM_015197
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr6_+_36853639
|
4.129
|
NM_152734
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr3_-_142166782
|
4.103
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr6_+_101846860
|
4.091
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr15_-_51914770
|
4.089
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr3_-_36986535
|
4.059
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr8_+_38034066
|
4.054
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr12_-_76953561
|
4.040
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr3_+_16974581
|
4.034
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chrX_+_70752911
|
4.022
|
NM_181672
NM_181673
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr6_+_34759743
|
3.987
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr2_-_48115846
|
3.977
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr6_-_91006460
|
3.922
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_+_70133179
|
3.876
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr4_-_48782281
|
3.842
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr10_+_181410
|
3.826
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr11_-_116968986
|
3.819
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr4_+_88896801
|
3.814
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_-_9009050
|
3.790
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr17_+_53342320
|
3.785
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr5_+_140165875
|
3.785
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr10_-_27149989
|
3.783
|
NM_001012750
NM_001012751
NM_001012752
NM_001178116
NM_001178119
NM_001178120
NM_001178121
NM_001178122
NM_001178123
NM_001178124
NM_001178125
NM_005470
|
ABI1
|
abl-interactor 1
|
|
chr2_+_64751436
|
3.755
|
NM_001002243
NM_017657
NM_203437
|
AFTPH
|
aftiphilin
|
|
chr6_-_88875766
|
3.746
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr5_+_71403040
|
3.733
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_+_90287404
|
3.720
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr19_-_36870064
|
3.696
|
NM_020917
|
ZFP14
|
zinc finger protein 14 homolog (mouse)
|
|
chr12_-_124457099
|
3.661
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr14_+_55738020
|
3.636
|
NM_017943
|
FBXO34
|
F-box protein 34
|
|
chr22_-_22221953
|
3.590
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr1_+_169075869
|
3.565
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr11_-_46939908
|
3.548
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr17_-_36956145
|
3.539
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr1_+_24286300
|
3.539
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr12_+_13197284
|
3.484
|
NM_020853
|
KIAA1467
|
KIAA1467
|
|
chr2_-_48132656
|
3.478
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr12_+_62654120
|
3.450
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr7_+_45614081
|
3.436
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr15_+_45879416
|
3.433
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr12_+_70172748
|
3.432
|
NM_001024647
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr3_-_50378238
|
3.414
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr5_+_140306301
|
3.413
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr10_-_3827418
|
3.393
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr12_+_70132562
|
3.377
|
NM_022456
NM_175624
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr4_-_141677470
|
3.372
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr20_+_11898536
|
3.358
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr17_+_61699786
|
3.351
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr20_+_37101457
|
3.345
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr7_+_66205642
|
3.345
|
NM_014504
|
RABGEF1
|
RAB guanine nucleotide exchange factor (GEF) 1
|
|
chr4_+_39063851
|
3.336
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr9_-_130742803
|
3.307
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr3_-_50374875
|
3.257
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr6_-_91006580
|
3.241
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr5_-_171615320
|
3.214
|
NM_005990
|
STK10
|
serine/threonine kinase 10
|