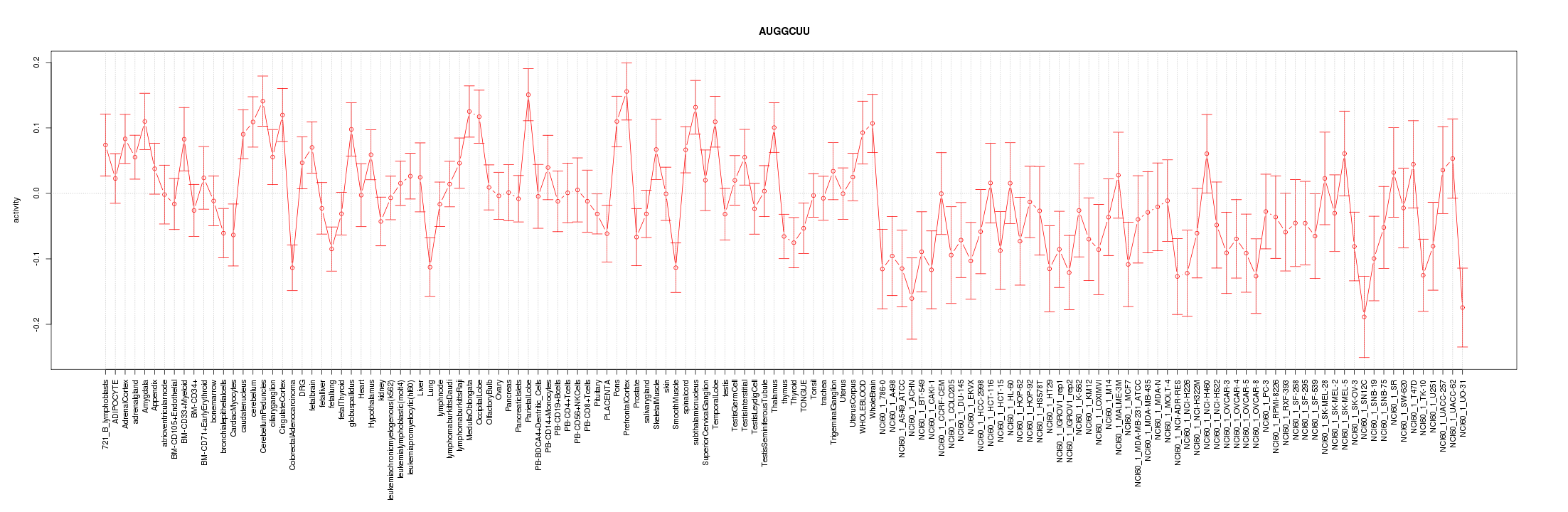
|
chr8_+_24771268
|
37.941
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_24772454
|
36.375
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr16_+_58498725
|
31.169
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58534045
|
30.678
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58497993
|
28.213
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58497548
|
26.816
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr1_+_171810611
|
22.040
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr12_+_79257772
|
21.931
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr2_+_131674223
|
20.939
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr12_+_79439432
|
19.725
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr16_+_8768403
|
19.261
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr5_-_88179278
|
19.249
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr12_+_79258448
|
19.236
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr9_-_93405066
|
18.958
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr3_+_49591897
|
18.394
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr15_+_91643429
|
18.177
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr11_-_117186918
|
17.220
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr9_-_79520878
|
17.115
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr15_+_91643181
|
17.040
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_27115897
|
16.854
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr17_+_40834571
|
16.310
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr17_-_10101854
|
16.197
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr5_-_136834887
|
15.952
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_+_121322848
|
15.642
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr19_-_38720312
|
15.550
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr17_-_9862765
|
15.059
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr11_-_117166247
|
14.447
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr16_+_8806825
|
14.248
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr17_-_8066254
|
14.195
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr4_-_87028805
|
13.257
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr17_-_78450247
|
13.182
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr16_+_8814567
|
12.068
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr17_-_9929567
|
11.974
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr7_+_45614081
|
11.851
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr19_-_17799005
|
11.762
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr5_-_88199868
|
11.320
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr20_-_32031425
|
11.195
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr8_+_38034066
|
11.162
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr1_-_155948317
|
11.087
|
NM_001162383
NM_001162384
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr1_+_183605207
|
10.997
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr10_-_62149487
|
10.940
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr4_-_87374282
|
10.876
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr18_+_32558207
|
10.768
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_-_155947898
|
10.735
|
NM_004723
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr10_-_62493282
|
10.413
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr17_-_9940004
|
10.382
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr2_+_166150340
|
10.223
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr14_+_69865380
|
9.919
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr10_-_81205091
|
9.909
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr8_-_18871163
|
9.624
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr19_+_18263998
|
9.573
|
NM_005027
|
PIK3R2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta)
|
|
chr4_-_87281374
|
9.256
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr6_+_138483024
|
9.178
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr3_-_10547267
|
9.138
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr1_-_202612580
|
9.089
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr19_+_10541334
|
8.969
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_-_38714781
|
8.635
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr7_+_149535508
|
8.605
|
NM_001099220
|
ZNF862
|
zinc finger protein 862
|
|
chr1_-_202679415
|
8.415
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr17_-_66287256
|
8.248
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr18_+_32556891
|
8.235
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr2_+_166152282
|
8.167
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr2_-_240322620
|
8.155
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr8_-_18666295
|
8.104
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_+_154074269
|
8.086
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr8_+_28747885
|
8.067
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr4_+_154125589
|
8.038
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr15_-_50978983
|
8.029
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr8_-_125740650
|
8.000
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr2_+_74273449
|
7.905
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr5_+_112043188
|
7.825
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr10_-_91403630
|
7.794
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr4_+_4387982
|
7.786
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr9_-_10612722
|
7.672
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr14_-_45431121
|
7.663
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr10_-_62332666
|
7.638
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_40973049
|
7.618
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr11_-_1593149
|
7.527
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr2_-_213403280
|
7.477
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr19_-_40324747
|
7.300
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr5_+_149380168
|
7.287
|
NM_014983
|
HMGXB3
|
HMG box domain containing 3
|
|
chr8_+_28748030
|
7.118
|
NM_024567
|
HMBOX1
|
homeobox containing 1
|
|
chr1_+_24882565
|
6.886
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr8_+_58906968
|
6.855
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr9_-_123476578
|
6.842
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr16_+_1664640
|
6.791
|
NM_020825
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr22_+_50247480
|
6.716
|
NM_014838
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr3_+_138067444
|
6.629
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr2_-_2334887
|
6.538
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr14_+_56584854
|
6.468
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr6_-_150185370
|
6.453
|
NM_032832
|
LRP11
|
low density lipoprotein receptor-related protein 11
|
|
chr3_+_138066489
|
6.424
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr4_-_42658883
|
6.381
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr15_+_31619043
|
6.372
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr13_+_35516390
|
6.251
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr5_+_112073555
|
6.139
|
NM_000038
NM_001127510
|
APC
|
adenomatous polyposis coli
|
|
chr7_+_152456820
|
6.126
|
NM_001040135
NM_020445
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast)
|
|
chr1_+_209757044
|
6.059
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr1_+_159141376
|
5.951
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr6_+_72922513
|
5.925
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_-_61900659
|
5.911
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_+_94929082
|
5.858
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr2_-_11484699
|
5.850
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr1_+_169075869
|
5.830
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr3_+_53529030
|
5.817
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr19_-_12833461
|
5.804
|
NM_001136196
|
TNPO2
|
transportin 2
|
|
chr8_-_20112691
|
5.795
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr19_-_12832923
|
5.767
|
NM_013433
|
TNPO2
|
transportin 2
|
|
chr20_-_48532067
|
5.737
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr1_-_46598250
|
5.720
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr11_+_62475113
|
5.710
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chrX_-_47509993
|
5.703
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr10_+_112836784
|
5.583
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chr22_+_38004454
|
5.574
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr5_-_114598547
|
5.574
|
NM_005023
|
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr14_+_99947732
|
5.545
|
NM_001099402
|
CCNK
|
cyclin K
|
|
chr17_-_7197866
|
5.491
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr15_+_27111865
|
5.452
|
NM_000810
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr15_+_27112106
|
5.436
|
NM_001165037
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr3_+_123813527
|
5.424
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr4_+_140374960
|
5.347
|
NM_031296
|
RAB33B
|
RAB33B, member RAS oncogene family
|
|
chr19_+_10531128
|
5.271
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr13_+_98086315
|
5.243
|
NM_021033
|
RAP2A
|
RAP2A, member of RAS oncogene family
|
|
chr3_-_56502364
|
5.227
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr7_+_4721722
|
5.214
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr2_-_175547471
|
5.212
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr5_-_160975124
|
5.134
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr5_+_76506705
|
5.089
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr15_-_33360084
|
4.956
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr18_+_48556483
|
4.942
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chrX_-_107019001
|
4.904
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_+_116853118
|
4.798
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr15_+_42066631
|
4.766
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr10_+_101419169
|
4.731
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr3_+_9691116
|
4.691
|
NM_001077525
NM_001077526
NM_022485
|
MTMR14
|
myotubularin related protein 14
|
|
chrX_-_106960268
|
4.652
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_53192021
|
4.610
|
NM_024646
|
ZYG11B
|
zyg-11 homolog B (C. elegans)
|
|
chr8_-_66753968
|
4.566
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr20_+_42875905
|
4.561
|
NM_024034
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr17_-_28562953
|
4.547
|
NM_001045
|
SLC6A4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4
|
|
chr12_-_58026846
|
4.512
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr22_-_42017032
|
4.434
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr5_-_147162251
|
4.409
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr19_-_344778
|
4.350
|
NM_017550
|
MIER2
|
mesoderm induction early response 1, family member 2
|
|
chr11_+_13299260
|
4.318
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr13_-_41240607
|
4.239
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr11_-_85338313
|
4.223
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_-_12834628
|
4.190
|
NM_001136195
|
TNPO2
|
transportin 2
|
|
chr12_+_58138663
|
4.155
|
NM_005981
|
TSPAN31
|
tetraspanin 31
|
|
chr2_+_7057471
|
4.085
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr6_+_17393610
|
4.080
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chrX_-_53310748
|
4.069
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr6_-_150039391
|
4.061
|
NM_004690
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr6_+_72926144
|
3.994
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr18_+_32621299
|
3.985
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr18_-_29523087
|
3.968
|
NM_014939
|
TRAPPC8
|
trafficking protein particle complex 8
|
|
chr4_-_16900348
|
3.937
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr2_-_211341374
|
3.822
|
NM_001136574
NM_006055
NM_001136575
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial)
|
|
chr17_-_7832752
|
3.796
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr1_+_203595897
|
3.768
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr10_+_86088156
|
3.753
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr12_+_4382882
|
3.751
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr8_+_104512975
|
3.718
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr19_+_10543110
|
3.717
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr1_+_27719151
|
3.715
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr2_-_201936389
|
3.708
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr17_-_62340652
|
3.697
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr2_+_128848752
|
3.691
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr2_+_166095911
|
3.687
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr3_+_142443214
|
3.663
|
NM_001251845
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chrX_-_106959597
|
3.644
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_-_184724021
|
3.614
|
NM_025191
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr2_-_175351755
|
3.538
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr16_+_639668
|
3.446
|
NM_001172664
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr19_-_2702738
|
3.444
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr6_+_73076431
|
3.391
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_+_64845839
|
3.387
|
NM_013254
|
TBK1
|
TANK-binding kinase 1
|
|
chr9_-_80646150
|
3.361
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr20_-_41818464
|
3.334
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr19_-_46295785
|
3.324
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr7_-_139876718
|
3.323
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr13_+_111806038
|
3.320
|
NM_003899
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr8_-_143695812
|
3.290
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr3_+_124303505
|
3.270
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr20_-_48099178
|
3.224
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr8_-_17658385
|
3.165
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_-_17579729
|
3.149
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr19_+_10563581
|
3.078
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_+_61699786
|
3.064
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr13_-_77460432
|
3.053
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr9_-_100395961
|
3.051
|
NM_139246
|
TSTD2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2
|
|
chr15_-_64648359
|
3.049
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr3_-_15106806
|
3.034
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr14_-_50698090
|
2.961
|
NM_006939
|
SOS2
|
son of sevenless homolog 2 (Drosophila)
|
|
chr9_-_139922705
|
2.916
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_+_205012329
|
2.912
|
NM_005076
|
CNTN2
|
contactin 2 (axonal)
|
|
chr3_+_43328001
|
2.895
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chrX_+_122318095
|
2.895
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chrX_-_135863502
|
2.893
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr19_+_2328628
|
2.865
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|