|
chr2_+_216176678
|
64.286
|
NM_004044
|
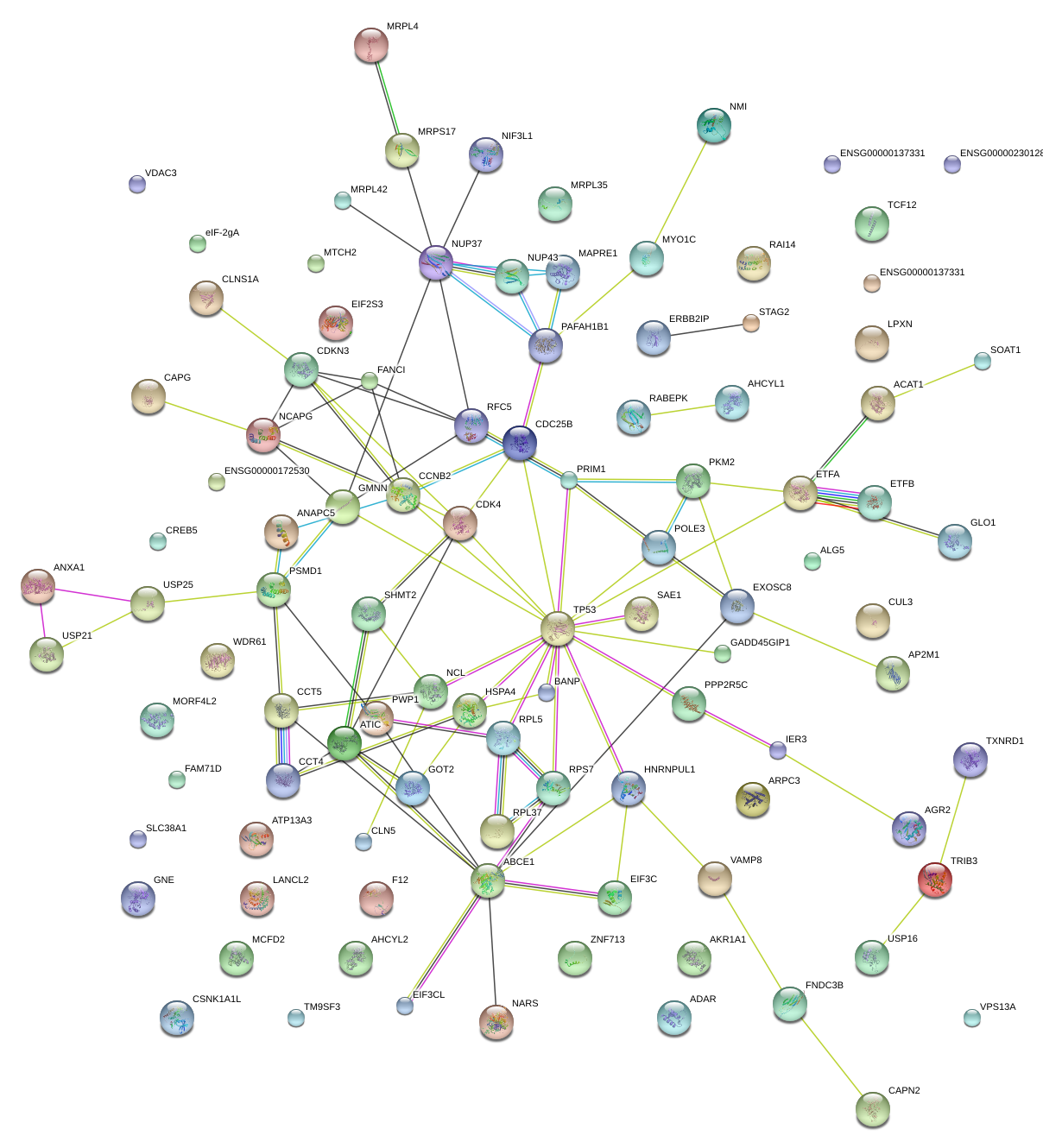
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr11_-_77348758
|
56.345
|
NM_001293
|
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr11_-_67450536
|
47.447
|
|
RPL37
|
ribosomal protein L37
|
|
chr2_+_216176808
|
44.223
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr15_-_72501017
|
43.772
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr15_-_76603806
|
42.914
|
NM_000126
NM_001127716
|
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide
|
|
chr14_+_67656109
|
40.658
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr5_+_132428394
|
39.286
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr21_+_30400229
|
37.346
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr10_-_98321823
|
34.794
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr7_+_56019552
|
34.721
|
NM_015969
|
MRPS17
|
mitochondrial ribosomal protein S17
|
|
chr13_+_37574916
|
34.417
|
|
EXOSC8
|
exosome component 8
|
|
chr6_-_30712294
|
33.860
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr12_-_58146118
|
33.721
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr15_-_76603791
|
33.659
|
|
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide
|
|
chr2_+_85804689
|
32.583
|
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chrX_+_123234462
|
32.509
|
|
STAG2
|
stromal antigen 2
|
|
chr9_+_75766794
|
32.227
|
|
ANXA1
|
annexin A1
|
|
chr14_+_102276125
|
32.221
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr10_-_98336445
|
30.816
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr12_+_104680712
|
30.736
|
NM_003330
NM_182729
NM_182742
NM_182743
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr9_+_75766777
|
30.328
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr17_-_1248346
|
30.278
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr16_+_28699878
|
29.902
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr1_-_154580632
|
29.469
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr12_-_121792011
|
28.506
|
NM_001137559
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr12_-_46633551
|
27.202
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr19_-_13068008
|
26.983
|
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr19_+_10362598
|
26.933
|
NM_015956
NM_146387
NM_146388
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr8_+_42249383
|
26.901
|
|
VDAC3
|
voltage-dependent anion channel 3
|
|
chr3_+_171757413
|
26.886
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr13_-_37679800
|
26.824
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr19_+_47634118
|
26.787
|
|
SAE1
|
SUMO1 activating enzyme subunit 1
|
|
chr13_+_37574677
|
26.475
|
NM_181503
|
EXOSC8
|
exosome component 8
|
|
chr2_-_152146373
|
26.265
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr12_-_57146081
|
26.243
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr19_+_47634044
|
26.090
|
NM_001145713
NM_001145714
NM_005500
|
SAE1
|
SUMO1 activating enzyme subunit 1
|
|
chr2_+_85804613
|
25.913
|
NM_003761
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chr11_-_47664014
|
25.607
|
NM_014342
|
MTCH2
|
mitochondrial carrier 2
|
|
chr12_-_110888152
|
25.238
|
NM_005719
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr15_+_59397276
|
25.216
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr15_+_57511616
|
25.207
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_+_223889253
|
25.124
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_93297614
|
25.022
|
|
RPL5
|
ribosomal protein L5
|
|
chr7_-_16844606
|
24.910
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr12_+_118454505
|
24.589
|
NM_001130112
NM_001206801
NM_007370
NM_181578
|
RFC5
|
replication factor C (activator 1) 5, 36.5kDa
|
|
chr9_-_36276930
|
24.466
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr12_+_93861265
|
24.321
|
NM_014050
NM_172177
|
MRPL42
|
mitochondrial ribosomal protein L42
|
|
chr20_+_361675
|
24.308
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr2_+_201756639
|
24.170
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr1_+_93297598
|
23.966
|
|
RPL5
|
ribosomal protein L5
|
|
chr2_+_216176818
|
23.903
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr1_+_46016524
|
23.752
|
|
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase)
|
|
chr1_+_46016454
|
23.589
|
NM_001202413
NM_001202414
NM_006066
NM_153326
|
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase)
|
|
chr20_+_3776385
|
23.439
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr6_-_150057659
|
22.802
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr2_-_47134945
|
22.734
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr5_-_176836488
|
22.028
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr2_+_86426555
|
21.909
|
NM_016622
NM_145644
|
MRPL35
|
mitochondrial ribosomal protein L35
|
|
chrX_-_102942968
|
21.893
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr2_-_232326394
|
21.891
|
|
NCL
|
nucleolin
|
|
chr12_+_108079516
|
21.864
|
NM_007062
|
PWP1
|
PWP1 homolog (S. cerevisiae)
|
|
chr1_+_93297532
|
21.720
|
NM_000969
|
RPL5
|
ribosomal protein L5
|
|
chr2_+_3622895
|
21.676
|
|
RPS7
|
ribosomal protein S7
|
|
chr5_+_65372744
|
21.522
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr11_-_58343308
|
21.410
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr19_+_41768368
|
21.395
|
NM_144732
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr12_-_102512277
|
21.349
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr6_+_24775154
|
21.250
|
NM_001251989
NM_001251990
NM_015895
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr16_-_58768132
|
21.215
|
NM_002080
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr2_+_3622909
|
21.058
|
|
RPS7
|
ribosomal protein S7
|
|
chr12_-_110888160
|
21.040
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr13_-_37573474
|
20.942
|
NM_001142364
NM_013338
|
ALG5
|
asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae)
|
|
chr6_-_38670829
|
20.610
|
|
GLO1
|
glyoxalase I
|
|
chr2_-_62115744
|
20.580
|
NM_006430
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr11_+_107992252
|
20.558
|
NM_000019
|
ACAT1
|
acetyl-CoA acetyltransferase 1
|
|
chr2_+_3622846
|
20.457
|
NM_001011
|
RPS7
|
ribosomal protein S7
|
|
chr9_+_79968344
|
20.435
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr19_-_13068033
|
20.426
|
NM_052850
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1
|
|
chr19_-_51858041
|
20.384
|
NM_001014763
|
ETFB
|
electron-transfer-flavoprotein, beta polypeptide
|
|
chrX_+_24073060
|
20.345
|
NM_001415
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa
|
|
chrX_+_24073071
|
20.063
|
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa
|
|
chr18_-_55289026
|
20.013
|
NM_004539
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr2_-_62115714
|
19.910
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr9_-_116172951
|
19.663
|
NM_017443
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr7_+_55433140
|
19.486
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr5_+_34687663
|
19.419
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr3_+_183892668
|
19.373
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr4_+_146031528
|
19.353
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr15_+_89787226
|
19.295
|
|
FANCI
|
Fanconi anemia, complementation group I
|
|
chr14_+_54863672
|
19.271
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr2_-_62115641
|
19.121
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr2_+_85804724
|
19.087
|
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chr12_+_57623355
|
19.036
|
NM_001166356
NM_005412
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr2_-_225370687
|
18.936
|
|
CUL3
|
cullin 3
|
|
chr20_+_31407696
|
18.825
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr5_+_10250440
|
18.723
|
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon)
|
|
chr17_-_7578810
|
18.709
|
NM_001126115
NM_001126116
NM_001126117
|
TP53
|
tumor protein p53
|
|
chr9_+_127962850
|
18.598
|
NM_005833
|
RABEPK
|
Rab9 effector protein with kelch motifs
|
|
chr16_+_88003623
|
18.539
|
NM_001173543
|
BANP
|
BTG3 associated nuclear protein
|
|
chr2_-_85641153
|
18.410
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr5_+_10250403
|
18.364
|
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon)
|
|
chr16_-_28437663
|
18.346
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr15_-_78591902
|
18.311
|
NM_025234
|
WDR61
|
WD repeat domain 61
|
|
chr3_+_183892699
|
18.305
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr19_+_41798147
|
18.264
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr1_+_110543548
|
18.223
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr10_+_102123420
|
18.216
|
|
|
|
|
chr2_-_85637486
|
18.016
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr2_+_231921562
|
17.922
|
NM_001191037
NM_002807
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr7_+_28725597
|
17.856
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_194188967
|
17.796
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr11_+_17332454
|
17.677
|
|
|
|
|
chr18_+_29671817
|
17.590
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr13_-_46756295
|
17.498
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr2_+_231921625
|
17.432
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr2_+_231921608
|
17.399
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr2_-_85641128
|
17.157
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr9_-_88715024
|
17.138
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr7_-_99699426
|
16.990
|
NM_005916
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr1_-_150207025
|
16.901
|
NM_001136479
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr18_-_55289014
|
16.836
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr7_-_66057331
|
16.784
|
|
LOC493754
|
RAB guanine nucleotide exchange factor (GEF) 1 pseudogene
|
|
chr2_-_62115726
|
16.762
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr19_+_13134782
|
16.744
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_123706393
|
16.637
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr9_+_127962809
|
16.616
|
NM_001174152
NM_001174153
|
RABEPK
|
Rab9 effector protein with kelch motifs
|
|
chr3_+_183892662
|
16.495
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr1_+_203830723
|
16.485
|
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr5_+_10250370
|
16.351
|
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon)
|
|
chr3_+_183892647
|
16.327
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chrX_-_129272014
|
16.148
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr7_-_87849341
|
16.114
|
|
SRI
|
sorcin
|
|
chr10_-_58121019
|
16.061
|
NM_001005413
NM_007057
NM_032997
|
ZWINT
|
ZW10 interactor
|
|
chr7_+_55433396
|
16.054
|
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr3_-_52569021
|
15.994
|
NM_022908
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr3_-_79817058
|
15.985
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr6_+_44187241
|
15.934
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr11_-_16996559
|
15.915
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr6_-_38670915
|
15.879
|
|
GLO1
|
glyoxalase I
|
|
chr14_-_55650348
|
15.875
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr6_-_38670882
|
15.813
|
|
GLO1
|
glyoxalase I
|
|
chr15_+_89787184
|
15.807
|
NM_001113378
NM_018193
|
FANCI
|
Fanconi anemia, complementation group I
|
|
chr16_+_56817367
|
15.803
|
NM_001242796
|
NUP93
|
nucleoporin 93kDa
|
|
chr3_-_49066813
|
15.743
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr1_+_203830732
|
15.693
|
NM_003094
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr10_-_58121011
|
15.514
|
|
ZWINT
|
ZW10 interactor
|
|
chr22_-_42343079
|
15.483
|
NM_001002876
NM_024053
|
CENPM
|
centromere protein M
|
|
chr17_+_4699456
|
15.460
|
NM_002798
|
PSMB6
|
proteasome (prosome, macropain) subunit, beta type, 6
|
|
chr6_+_34725266
|
15.439
|
|
SNRPC
|
small nuclear ribonucleoprotein polypeptide C
|
|
chr12_+_16064105
|
15.372
|
NM_015954
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr2_+_171784947
|
15.369
|
NM_001201428
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr10_-_98325100
|
15.328
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr12_-_120638912
|
15.307
|
NM_001002
NM_053275
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr1_-_45452266
|
15.161
|
NM_001166588
NM_020365
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr5_+_10250410
|
15.114
|
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon)
|
|
chr2_+_170662044
|
14.973
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr9_-_35079751
|
14.863
|
NM_004629
|
FANCG
|
Fanconi anemia, complementation group G
|
|
chr7_+_94292737
|
14.803
|
|
PEG10
|
paternally expressed 10
|
|
chr1_+_203830763
|
14.759
|
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr18_+_29671925
|
14.751
|
|
RNF138
|
ring finger protein 138
|
|
chr12_+_16064232
|
14.683
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr13_-_46756326
|
14.602
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr8_-_71581419
|
14.538
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr12_+_16064282
|
14.533
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr2_+_223725675
|
14.446
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr2_+_231921595
|
14.373
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr6_+_119215240
|
14.341
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr9_-_130700762
|
14.339
|
NM_003863
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit
|
|
chr9_+_110047049
|
14.331
|
NM_001244713
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr3_-_49066858
|
14.327
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr12_-_96429397
|
14.288
|
|
LTA4H
|
leukotriene A4 hydrolase
|
|
chr17_+_4699473
|
14.030
|
|
PSMB6
|
proteasome (prosome, macropain) subunit, beta type, 6
|
|
chr6_-_170862326
|
14.014
|
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1
|
|
chr6_-_38670871
|
13.995
|
|
GLO1
|
glyoxalase I
|
|
chr18_+_3447607
|
13.921
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_154580481
|
13.848
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr12_+_56915587
|
13.837
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr1_-_93646245
|
13.815
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr16_-_47005339
|
13.744
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr7_+_28475233
|
13.710
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_32996961
|
13.692
|
NM_007270
|
FKBP9
|
FK506 binding protein 9, 63 kDa
|
|
chr3_-_185655806
|
13.682
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr7_+_94292816
|
13.670
|
|
PEG10
|
paternally expressed 10
|
|
chr8_-_71520433
|
13.647
|
|
TRAM1
|
translocation associated membrane protein 1
|
|
chr7_-_95064213
|
13.566
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr2_-_18770776
|
13.553
|
NM_001199103
NM_001199104
NM_001002006
NM_001199086
NM_001199087
NM_001199088
NM_033253
|
NT5C1B-RDH14
NT5C1B
|
NT5C1B-RDH14 readthrough
5'-nucleotidase, cytosolic IB
|
|
chr12_+_64803741
|
13.459
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr10_-_27057776
|
13.338
|
|
ABI1
|
abl-interactor 1
|
|
chr3_+_183892569
|
13.335
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr19_+_3198813
|
13.229
|
|
NCLN
|
nicalin
|
|
chr9_-_14086901
|
13.197
|
|
NFIB
|
nuclear factor I/B
|
|
chr6_-_24719286
|
13.068
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr9_+_110046333
|
12.978
|
NM_001244724
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr12_+_32832128
|
12.912
|
NM_005690
NM_012062
NM_012063
|
DNM1L
|
dynamin 1-like
|
|
chr12_-_120638909
|
12.857
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr5_+_162864571
|
12.761
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr6_-_38670904
|
12.755
|
|
GLO1
|
glyoxalase I
|
|
chr17_+_26794838
|
12.747
|
|
|
|
|
chr22_-_41252646
|
12.719
|
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein)
|