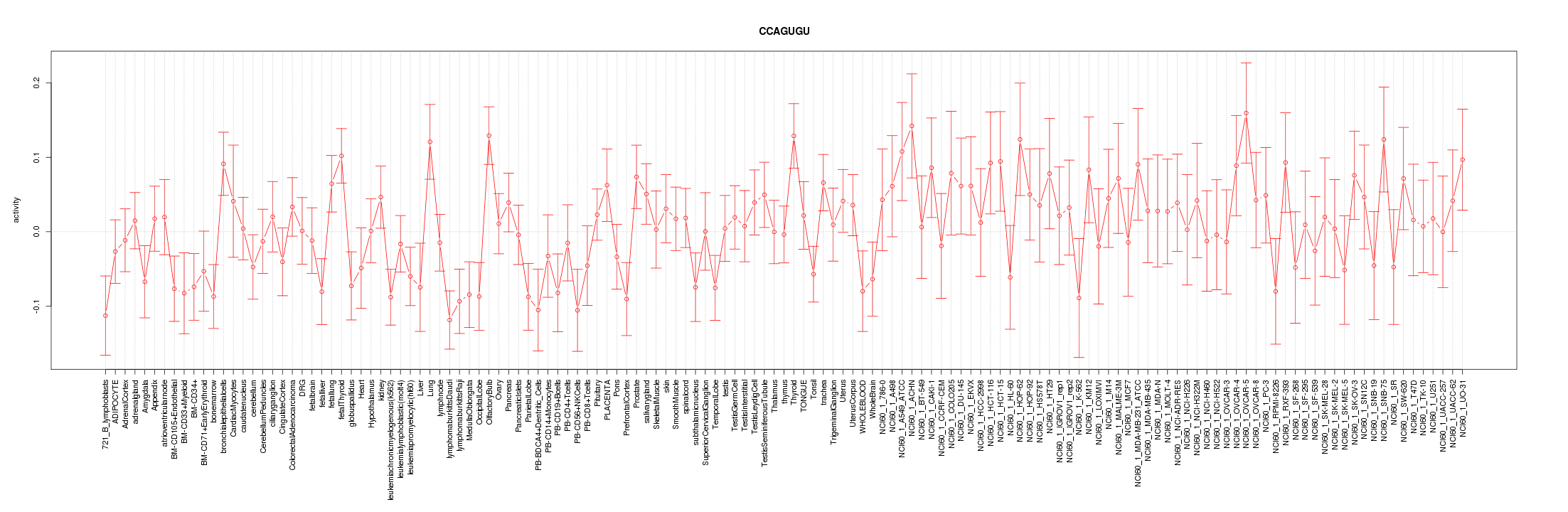
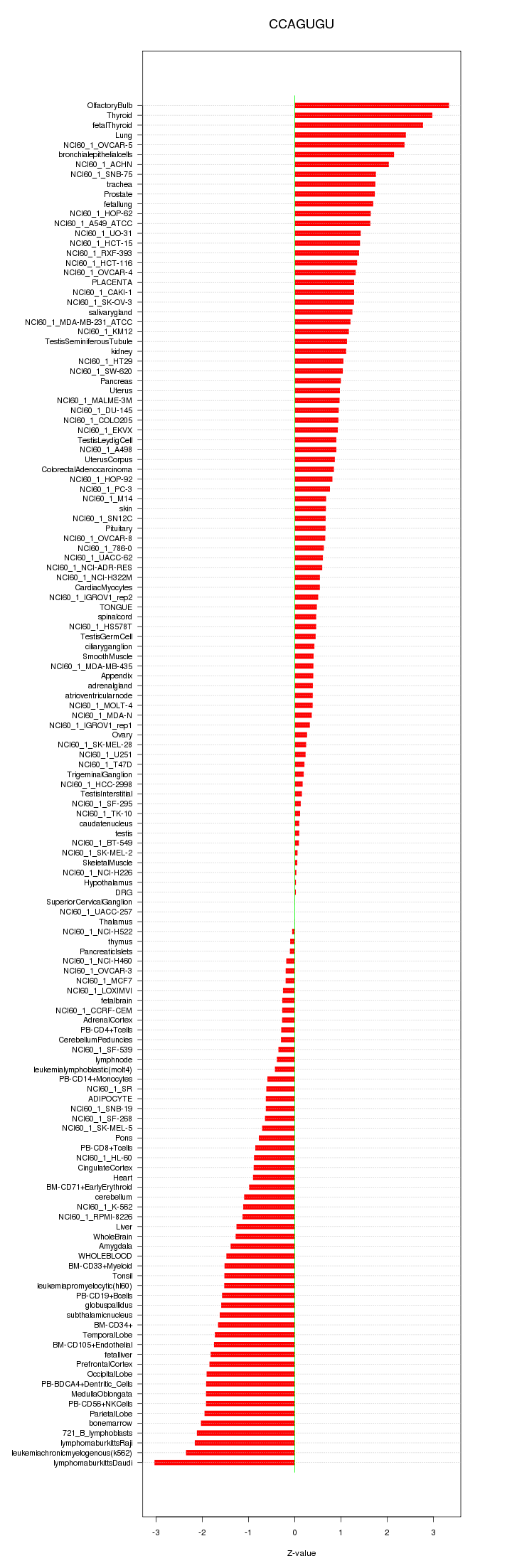
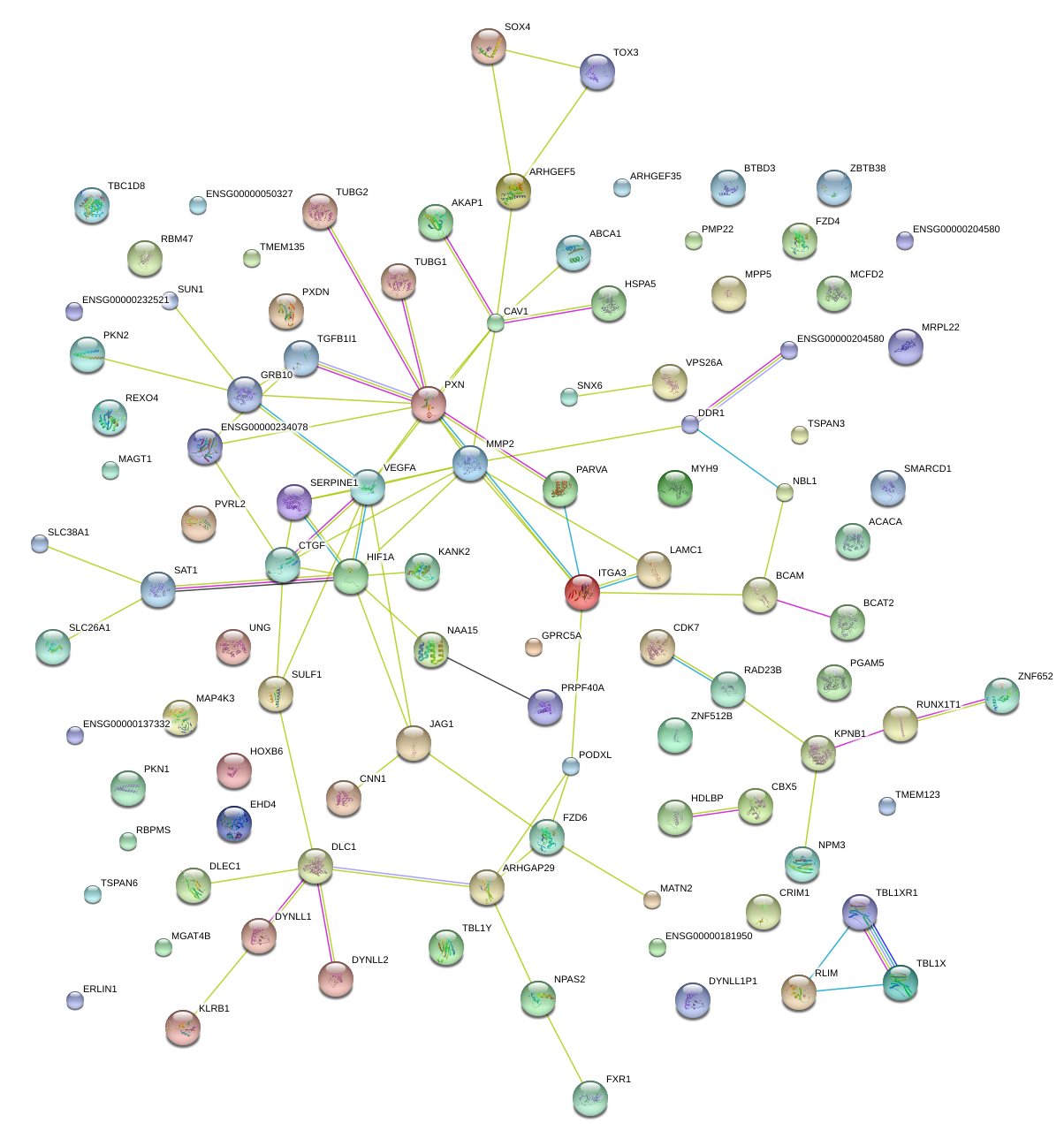
|
chr7_+_116166346
|
27.406
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116164838
|
26.999
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116165043
|
26.034
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chrX_-_99891690
|
17.544
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr1_-_94703252
|
15.588
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_182992329
|
14.033
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr17_+_48133331
|
13.508
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr12_+_13043955
|
12.630
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr8_+_104310660
|
12.246
|
NM_001164615
|
FZD6
|
frizzled family receptor 6
|
|
chr8_+_104311058
|
12.162
|
NM_001164616
NM_003506
|
FZD6
|
frizzled family receptor 6
|
|
chr7_+_100770378
|
11.827
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr6_+_30851860
|
11.546
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr7_-_131241321
|
10.455
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr6_+_30852326
|
10.375
|
NM_013993
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_+_30852756
|
10.252
|
NM_001202523
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_+_30856462
|
10.201
|
NM_001202521
NM_001202522
NM_013994
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr1_+_19969722
|
9.698
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_+_19972259
|
9.459
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr7_+_872137
|
9.318
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr1_+_19970207
|
8.869
|
NM_001204084
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_+_19971865
|
8.851
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr8_-_12973752
|
8.503
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr4_-_40517913
|
8.383
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr2_-_242255114
|
8.349
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr1_+_19970674
|
7.976
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr8_-_12990731
|
7.836
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_-_11308184
|
7.396
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr5_-_179233772
|
7.208
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr17_-_15165873
|
7.151
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_-_15168590
|
6.956
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr8_+_98881277
|
6.940
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr3_+_180630054
|
6.895
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr19_-_11305186
|
6.881
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr9_-_128003634
|
6.703
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr8_+_30241943
|
6.427
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr17_-_15164043
|
6.360
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_-_46682333
|
6.280
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr16_+_55515468
|
6.266
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr5_+_68530621
|
6.177
|
NM_001799
|
CDK7
|
cyclin-dependent kinase 7
|
|
chr2_+_36583369
|
6.116
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr5_-_179229874
|
6.068
|
NM_054013
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr15_-_77363519
|
6.006
|
NM_001168412
NM_005724
NM_198902
|
TSPAN3
|
tetraspanin 3
|
|
chr9_+_110045516
|
5.870
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr12_-_54653317
|
5.831
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr17_+_55163077
|
5.534
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_-_47143250
|
5.525
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr16_+_55512801
|
5.477
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr19_+_11649578
|
5.447
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr6_+_43737937
|
5.440
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_-_47142950
|
5.410
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr10_+_70883856
|
5.316
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr17_+_55162530
|
5.310
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr12_-_120687956
|
5.258
|
NM_025157
|
PXN
|
paxillin
|
|
chr14_+_62162042
|
5.251
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr12_-_54653369
|
5.236
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr2_-_242212244
|
5.221
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_-_120703549
|
5.167
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr4_-_40631846
|
5.079
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr2_-_47168902
|
4.987
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr8_-_93029864
|
4.790
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_153573858
|
4.760
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr20_-_10654429
|
4.724
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr20_-_62601140
|
4.585
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr6_+_21593963
|
4.574
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr11_-_102323353
|
4.529
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr7_-_50800018
|
4.286
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chrX_+_23801274
|
4.137
|
NM_002970
|
SAT1
|
spermidine/spermine N1-acetyltransferase 1
|
|
chr16_+_31483066
|
4.134
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr7_-_50772997
|
3.974
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr14_+_67707825
|
3.859
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr11_-_86666211
|
3.793
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr7_+_144052432
|
3.791
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr11_+_12399024
|
3.559
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr2_-_101767714
|
3.502
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr17_+_40761357
|
3.491
|
NM_001070
|
TUBG1
|
tubulin, gamma 1
|
|
chr17_-_47439326
|
3.465
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr1_+_19923457
|
3.375
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr9_-_107690526
|
3.336
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr7_-_50861114
|
3.317
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr16_+_31484497
|
3.286
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr5_+_154320628
|
3.285
|
NM_001014990
NM_014180
|
MRPL22
|
mitochondrial ribosomal protein L22
|
|
chr3_-_176915012
|
3.221
|
NM_024665
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr4_+_140222668
|
3.161
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr19_+_45312294
|
3.083
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr10_-_101945787
|
3.080
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr17_-_35656691
|
3.042
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr3_+_141043054
|
2.881
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr2_-_39664129
|
2.851
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr10_-_101945686
|
2.748
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr11_+_86748870
|
2.724
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr15_-_42264749
|
2.724
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chrX_-_73834460
|
2.658
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr19_+_45349392
|
2.653
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr22_-_36784055
|
2.574
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr20_+_11898536
|
2.560
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr1_+_89149836
|
2.527
|
NM_006256
|
PKN2
|
protein kinase N2
|
|
chrX_-_77150907
|
2.471
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr12_+_50478942
|
2.461
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr16_-_52581713
|
2.456
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_+_70405027
|
2.447
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr6_-_132272311
|
2.399
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr14_-_35099296
|
2.397
|
NM_021249
NM_152233
|
SNX6
|
sorting nexin 6
|
|
chr17_+_45727274
|
2.363
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr12_+_109535922
|
2.354
|
NM_003362
|
UNG
|
uracil-DNA glycosylase
|
|
chr2_+_101436491
|
2.333
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr12_+_109535377
|
2.307
|
NM_080911
|
UNG
|
uracil-DNA glycosylase
|
|
chr1_-_24127157
|
2.298
|
NM_001008216
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr10_-_98346793
|
2.279
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr11_+_9406123
|
2.251
|
NM_006391
|
IPO7
|
importin 7
|
|
chr17_-_39968450
|
2.232
|
NM_006455
|
LEPREL4
|
leprecan-like 4
|
|
chr2_+_241938254
|
2.203
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr17_-_35766762
|
2.198
|
NM_198834
NM_198839
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr11_+_34073188
|
2.178
|
NM_005898
NM_203364
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr12_+_52347163
|
2.158
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr19_+_39616384
|
2.130
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr12_-_122907083
|
2.124
|
NM_001247997
NM_002956
NM_198240
|
CLIP1
|
CAP-GLY domain containing linker protein 1
|
|
chr6_+_10555948
|
2.117
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr6_+_10585978
|
2.082
|
NM_145655
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr12_-_54673901
|
2.076
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr12_+_102271082
|
1.996
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr1_+_28764660
|
1.991
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr8_-_93107442
|
1.977
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_130794854
|
1.966
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr17_+_43224683
|
1.958
|
NM_006460
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr20_+_19870209
|
1.930
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_-_24126031
|
1.917
|
NM_001127621
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr12_+_50794591
|
1.907
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr3_-_189840225
|
1.894
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr17_+_35849950
|
1.884
|
NM_007026
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr12_+_104609501
|
1.866
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr12_+_104680712
|
1.844
|
NM_003330
NM_182729
NM_182742
NM_182743
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr5_-_83016850
|
1.836
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr8_+_110346549
|
1.832
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr1_+_166808721
|
1.828
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr17_+_46125675
|
1.819
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr17_-_4269814
|
1.801
|
NM_003342
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1
|
|
chr3_-_183273407
|
1.795
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr16_-_71842892
|
1.783
|
NM_001030007
NM_001128
|
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit
|
|
chr20_+_19867075
|
1.759
|
NM_001242581
|
RIN2
|
Ras and Rab interactor 2
|
|
chr13_-_24007822
|
1.748
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr3_-_189838907
|
1.668
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr3_-_160283361
|
1.621
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr14_+_92788924
|
1.620
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr1_+_13910251
|
1.604
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr5_+_179921371
|
1.601
|
NM_015455
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6
|
|
chr11_-_27528298
|
1.597
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chr7_+_856251
|
1.575
|
NM_001171944
NM_001171945
NM_025154
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr1_-_24126891
|
1.559
|
NM_000403
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr2_-_70781146
|
1.556
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr2_+_70485204
|
1.546
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr16_-_30798206
|
1.546
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr1_+_28696056
|
1.542
|
NM_001048183
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr2_+_109335906
|
1.537
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr21_+_40177847
|
1.536
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr5_+_149887673
|
1.532
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr3_-_125094003
|
1.532
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr10_-_71930185
|
1.526
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr5_-_176778683
|
1.524
|
NM_006816
|
LMAN2
|
lectin, mannose-binding 2
|
|
chr15_-_34394016
|
1.494
|
NM_020154
|
C15orf24
|
chromosome 15 open reading frame 24
|
|
chr1_-_205719340
|
1.471
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr11_-_57298180
|
1.445
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr18_-_54305839
|
1.439
|
NM_004786
|
TXNL1
|
thioredoxin-like 1
|
|
chr12_+_72148653
|
1.432
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr1_+_212788359
|
1.429
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_212738675
|
1.428
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_218518675
|
1.396
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr8_-_77913279
|
1.388
|
NM_001172086
NM_001172087
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr1_+_13911966
|
1.373
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr3_-_195270004
|
1.359
|
NM_006241
|
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2
|
|
chr2_+_170590241
|
1.358
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr11_-_62414096
|
1.337
|
NM_198334
NM_198335
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr12_-_46766563
|
1.329
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr8_-_77912296
|
1.315
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr7_-_105162551
|
1.309
|
NM_019042
|
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae)
|
|
chr1_-_214724974
|
1.304
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr16_+_16043206
|
1.300
|
NM_004996
NM_019862
NM_019898
NM_019899
NM_019900
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chrX_+_40944858
|
1.297
|
NM_001039590
NM_001039591
|
USP9X
|
ubiquitin specific peptidase 9, X-linked
|
|
chrX_-_77395108
|
1.296
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr20_-_24973410
|
1.279
|
NM_020531
|
C20orf3
|
chromosome 20 open reading frame 3
|
|
chr12_-_56652118
|
1.239
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr1_+_212781969
|
1.235
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_145826928
|
1.201
|
NM_001097612
NM_001097613
|
GPR89B
GPR89A
|
G protein-coupled receptor 89B
G protein-coupled receptor 89A
|
|
chr9_-_127952037
|
1.192
|
NM_001123355
NM_001123369
NM_002721
|
PPP6C
|
protein phosphatase 6, catalytic subunit
|
|
chr20_+_11871364
|
1.188
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr2_+_178257371
|
1.177
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr20_-_25566137
|
1.169
|
NM_025176
|
NINL
|
ninein-like
|
|
chr19_+_11925077
|
1.144
|
NM_152357
|
ZNF440
|
zinc finger protein 440
|
|
chr7_-_134855530
|
1.143
|
NM_001243749
NM_001243751
NM_001243754
NM_024033
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr9_-_94186140
|
1.137
|
NM_005384
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr17_-_56065596
|
1.090
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chrX_+_128674181
|
1.076
|
NM_000276
NM_001587
|
OCRL
|
oculocerebrorenal syndrome of Lowe
|
|
chr1_-_155881163
|
1.074
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr16_+_57673206
|
1.063
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr8_-_102217868
|
1.056
|
NM_001042511
|
ZNF706
|
zinc finger protein 706
|
|
chr8_-_102217907
|
1.032
|
NM_001042510
NM_016096
|
ZNF706
|
zinc finger protein 706
|
|
chr20_-_48184652
|
1.029
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr3_+_63884074
|
1.027
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr22_-_18507151
|
1.022
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr12_-_4754275
|
1.010
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr3_+_50192847
|
1.006
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|