|
chr15_+_31619043
|
38.547
|
NM_015995
|
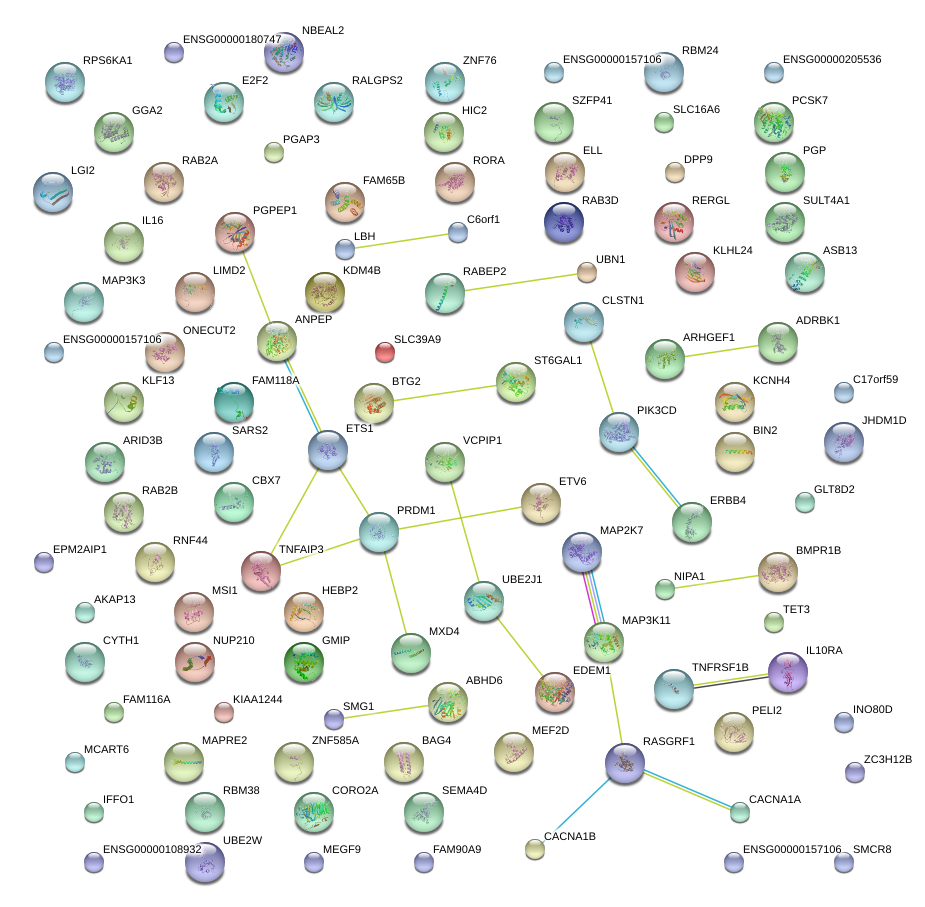
KLF13
|
Kruppel-like factor 13
|
|
chr6_+_138188352
|
28.764
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr20_+_55966440
|
24.322
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr16_-_28936454
|
23.963
|
NM_024816
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr22_-_39548448
|
23.746
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chrX_+_64708614
|
23.582
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr4_-_2263706
|
23.410
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr3_-_57678594
|
22.351
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr2_+_30454396
|
21.877
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr17_-_61777478
|
21.509
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr15_-_61521478
|
20.815
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr14_+_69865380
|
20.568
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr2_-_206950895
|
20.522
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr3_+_186648273
|
20.234
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr3_+_186739643
|
20.137
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr22_-_44258210
|
19.898
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr19_-_18632886
|
19.617
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr8_+_38034066
|
19.445
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr19_+_7968764
|
18.747
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_+_42387249
|
18.388
|
NM_004706
NM_198977
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr17_+_18218569
|
17.924
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr17_-_76778306
|
16.916
|
NM_004762
NM_017456
|
CYTH1
|
cytohesin 1
|
|
chr15_-_60884615
|
16.892
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_-_6665220
|
16.832
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr17_-_66287256
|
16.644
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr15_-_90358057
|
16.513
|
NM_001150
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr19_+_4969123
|
16.505
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr12_-_120806893
|
16.417
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr1_+_26856240
|
16.323
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr17_-_8093492
|
16.290
|
NM_017622
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr19_+_42388445
|
16.220
|
NM_199002
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr15_+_81589240
|
16.194
|
NM_004513
|
IL16
|
interleukin 16
|
|
chr6_-_24911194
|
16.111
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr15_+_74833517
|
16.083
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr1_-_156470506
|
15.999
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr11_+_117857062
|
15.949
|
NM_001558
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr16_-_2264776
|
15.728
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr17_+_61699786
|
15.718
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr9_-_123476578
|
15.714
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr19_-_4723814
|
15.471
|
NM_139159
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr3_+_5229357
|
15.424
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr15_-_79383080
|
15.380
|
NM_001145648
NM_002891
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr6_+_138483024
|
15.339
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr22_+_45705766
|
15.261
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr15_-_23086290
|
15.088
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr12_-_51717905
|
14.961
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr11_-_128392061
|
14.937
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr3_+_58223228
|
14.923
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr1_+_12227050
|
14.859
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr7_-_139876718
|
14.848
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr4_+_95679075
|
14.839
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr11_-_128457452
|
14.822
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr11_-_117102810
|
14.821
|
NM_004716
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chrX_-_103401607
|
14.662
|
NM_001012755
|
MCART6
|
mitochondrial carrier triple repeat 6
|
|
chr10_-_5708542
|
14.280
|
NM_024701
|
ASB13
|
ankyrin repeat and SOCS box containing 13
|
|
chr19_-_19754452
|
14.090
|
NM_016573
|
GMIP
|
GEM interacting protein
|
|
chr3_+_183353355
|
14.067
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr11_+_67033889
|
13.859
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr2_+_74273449
|
13.844
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr9_-_92094610
|
13.757
|
NM_001142287
NM_006378
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr6_+_106546736
|
13.592
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr3_+_47021146
|
13.369
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr1_-_23857711
|
13.307
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr15_+_85923739
|
13.182
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr16_+_4897911
|
13.180
|
NM_001079514
|
UBN1
|
ubinuclein 1
|
|
chr1_+_9711781
|
13.132
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr4_-_25032306
|
12.990
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr17_-_40333190
|
12.784
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr1_+_203274646
|
12.680
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr18_+_55102777
|
12.630
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr8_-_67579446
|
12.196
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr2_-_213403280
|
12.193
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr14_+_56584854
|
12.147
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr15_+_81517639
|
12.134
|
NM_172217
|
IL16
|
interleukin 16
|
|
chr9_-_100954883
|
12.108
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr9_+_140772228
|
11.977
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr15_+_81489218
|
11.934
|
NM_001172128
|
IL16
|
interleukin 16
|
|
chr19_-_37701385
|
11.798
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr15_-_77712436
|
11.692
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr8_-_74791098
|
11.662
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr6_+_35227478
|
11.638
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr3_-_13461802
|
11.531
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr12_+_11802726
|
11.355
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr22_+_21771661
|
11.338
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr19_-_11450254
|
11.323
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr18_+_32558207
|
11.276
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr14_-_21945031
|
11.247
|
NM_032846
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr17_-_37844266
|
11.237
|
NM_033419
|
PGAP3
|
post-GPI attachment to proteins 3
|
|
chr11_-_65381641
|
11.226
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr5_-_175964223
|
11.164
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr15_-_60919642
|
11.133
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_-_23521803
|
11.037
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr1_+_178694273
|
10.959
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr14_-_21944826
|
10.826
|
NM_001163380
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr6_-_90062610
|
10.707
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr6_+_106534188
|
10.695
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr16_-_18937722
|
10.691
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr2_-_99347588
|
10.690
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_-_211307314
|
10.613
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr6_+_11538486
|
10.364
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_-_202679415
|
10.058
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chrX_-_131623994
|
10.037
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr8_-_66753968
|
9.931
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr17_-_46651809
|
9.834
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr15_-_70390229
|
9.818
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr14_-_88789531
|
9.801
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr22_+_45704821
|
9.777
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr18_-_43652153
|
9.776
|
NM_024430
|
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2
|
|
chr14_+_70078309
|
9.733
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr8_-_41754244
|
9.682
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr2_-_74710356
|
9.626
|
NM_032779
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr22_+_41777932
|
9.394
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr1_+_182758583
|
9.385
|
NM_001200050
NM_001200051
NM_001200052
NM_001200056
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr4_+_75858205
|
9.369
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr16_+_4902076
|
9.361
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr13_-_73356070
|
9.336
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr18_+_48556483
|
9.323
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr7_-_143059696
|
9.297
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr6_+_391738
|
9.283
|
NM_001195286
NM_002460
|
IRF4
|
interferon regulatory factor 4
|
|
chr11_+_63606346
|
9.102
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr1_+_151584514
|
9.100
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr18_+_32556891
|
9.013
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_-_119234859
|
8.973
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr5_+_56110899
|
8.871
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr7_-_143059144
|
8.621
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr2_+_131113579
|
8.593
|
NM_001142370
NM_014369
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr5_-_38556682
|
8.560
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr5_+_133861685
|
8.474
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr3_+_49591897
|
8.468
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr18_+_32621299
|
8.456
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr5_-_159739482
|
8.430
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr15_+_86220267
|
8.303
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr7_+_2557496
|
8.294
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr19_-_5340700
|
8.267
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr3_-_50378238
|
8.255
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr9_-_72287097
|
8.223
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr1_+_78245306
|
8.146
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr11_+_1411089
|
8.067
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr20_+_35201948
|
7.958
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr5_-_37839781
|
7.942
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr4_+_72052354
|
7.922
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr11_+_71639767
|
7.917
|
NM_018320
|
RNF121
|
ring finger protein 121
|
|
chr15_-_73661604
|
7.887
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chrX_+_153639868
|
7.868
|
NM_000116
NM_181311
NM_181312
NM_181313
|
TAZ
|
tafazzin
|
|
chr17_-_36956145
|
7.863
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr1_-_200992824
|
7.855
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr10_-_103879933
|
7.810
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr1_-_212208779
|
7.771
|
NM_001199809
NM_001199811
NM_001199812
NM_015434
|
INTS7
|
integrator complex subunit 7
|
|
chr1_+_54519259
|
7.693
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr11_+_94277005
|
7.680
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr4_+_37455551
|
7.679
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr16_+_27561441
|
7.654
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr1_-_21616888
|
7.544
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr19_+_18794391
|
7.500
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr4_+_7045142
|
7.440
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr19_-_17799005
|
7.371
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr11_+_75526211
|
7.341
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr20_+_34359904
|
7.336
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr14_-_92572950
|
7.332
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr19_-_4066739
|
7.241
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr12_+_51632507
|
7.194
|
NM_001136264
NM_001136266
NM_001136267
NM_001136268
NM_001136269
NM_014764
|
DAZAP2
|
DAZ associated protein 2
|
|
chr12_-_42538432
|
7.107
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr12_-_123380682
|
7.087
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr7_-_98741681
|
7.064
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr13_-_29292961
|
7.053
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr3_-_52090090
|
7.044
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr12_+_53773924
|
7.032
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr3_+_9773427
|
7.012
|
NM_001003694
NM_004634
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr3_-_50374875
|
6.978
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr9_-_115095933
|
6.974
|
NM_001163788
NM_001163790
NM_001244896
NM_001244897
NM_005156
|
PTBP3
|
polypyrimidine tract binding protein 3
|
|
chr3_+_185000714
|
6.919
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr1_-_150552127
|
6.899
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr1_+_205197031
|
6.886
|
NM_014858
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr14_-_90085325
|
6.879
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr3_-_133748828
|
6.806
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr3_+_15469052
|
6.799
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr2_-_201936389
|
6.778
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr19_+_13261272
|
6.748
|
NM_004907
|
IER2
|
immediate early response 2
|
|
chr1_-_108507474
|
6.729
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_170626425
|
6.680
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_+_46923738
|
6.642
|
NM_001184744
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr20_+_35201854
|
6.624
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr20_+_54933982
|
6.620
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr17_+_21279667
|
6.618
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr14_+_102228134
|
6.595
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr9_-_99382073
|
6.552
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr14_-_82000198
|
6.527
|
NM_001244984
NM_005065
|
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr4_+_38665587
|
6.515
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr2_+_61292999
|
6.513
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr12_+_51818592
|
6.414
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr18_-_60986508
|
6.385
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr21_-_40685697
|
6.384
|
NM_001007246
NM_018963
NM_033656
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr1_-_20834609
|
6.377
|
NM_024544
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr1_-_173176314
|
6.376
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr16_+_68298326
|
6.329
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr5_+_96294154
|
6.320
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr12_+_5019048
|
6.317
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr17_-_27224708
|
6.311
|
NM_004475
|
FLOT2
|
flotillin 2
|
|
chr19_-_2051222
|
6.303
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr3_-_49823972
|
6.297
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|