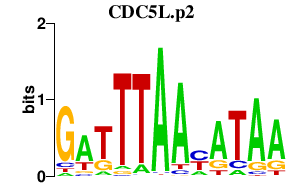
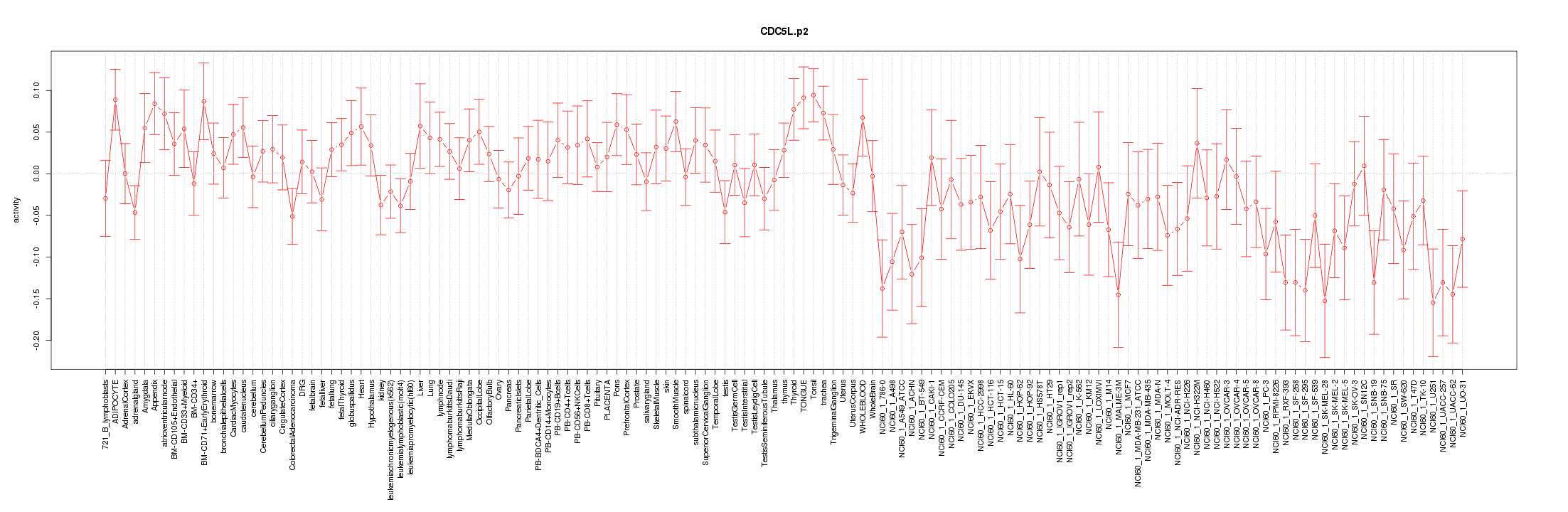
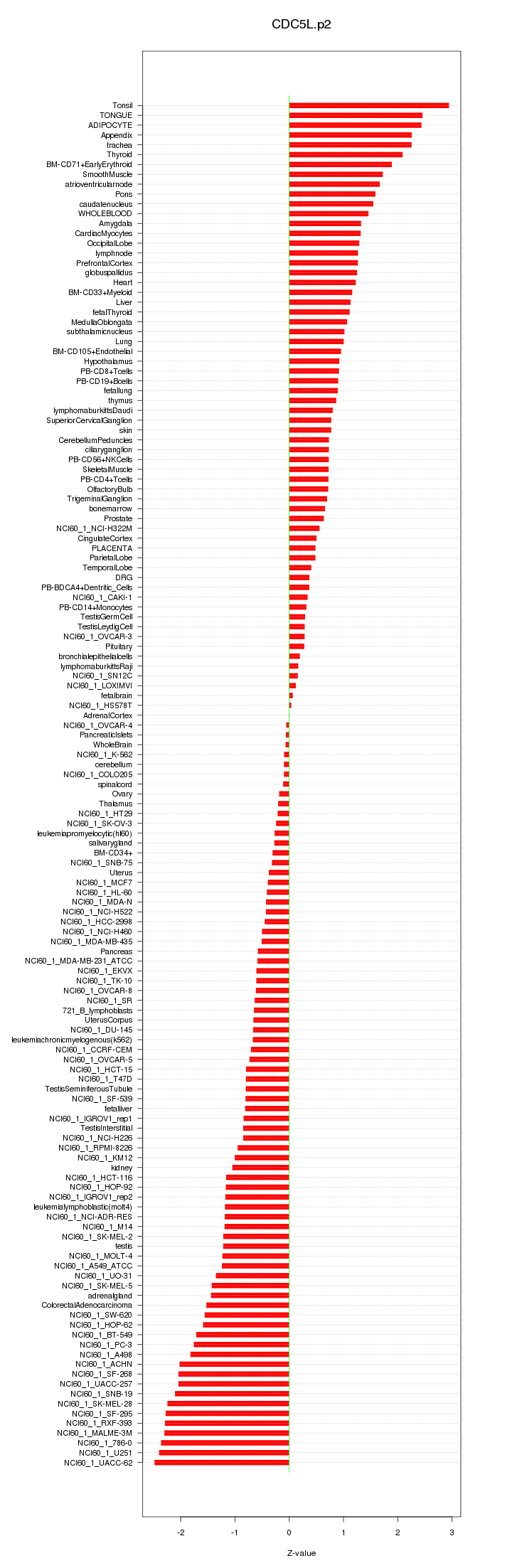
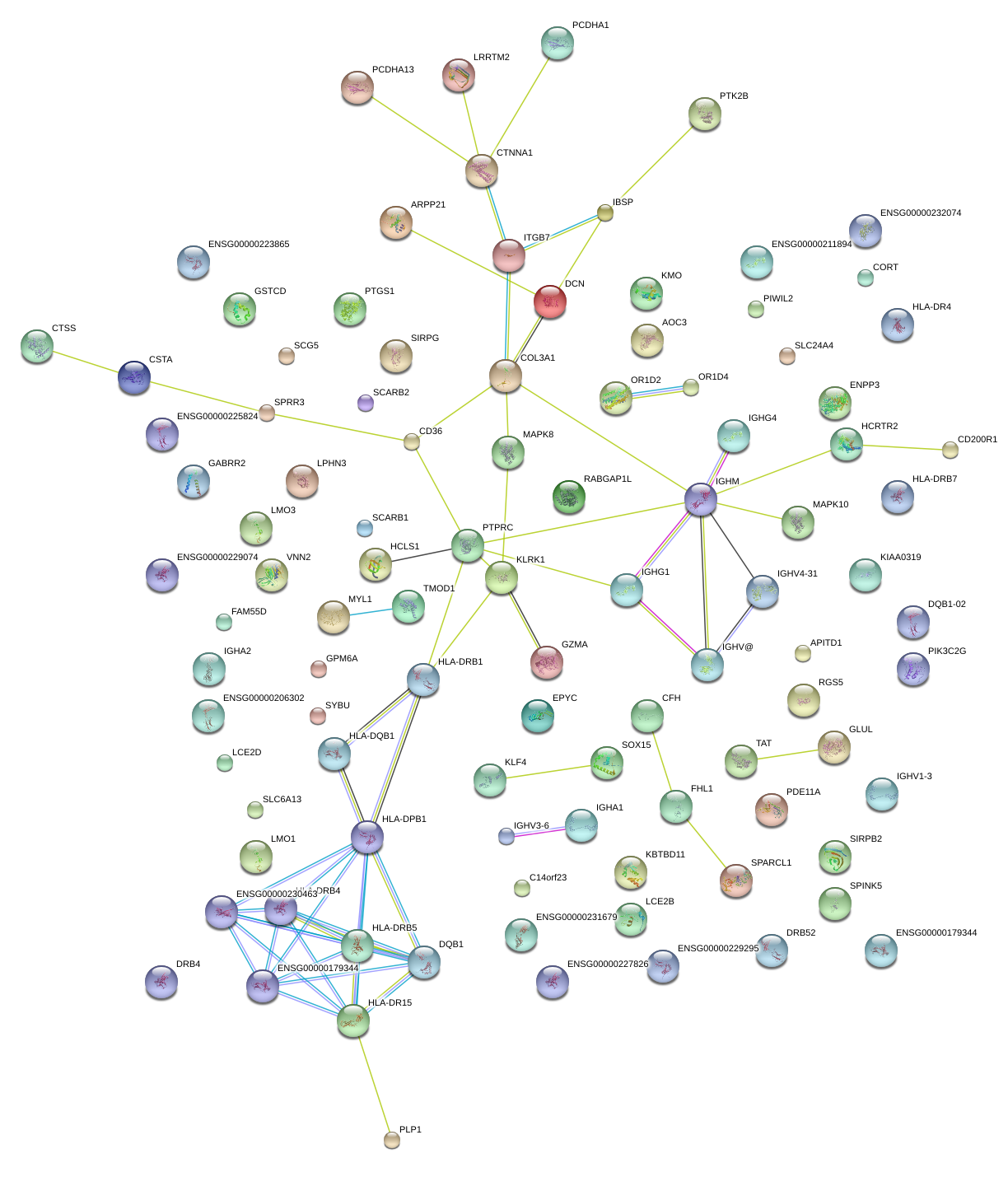
|
chr19_-_21646685
|
18.935
|
|
|
|
|
chr5_-_138210992
|
18.278
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr16_-_71610948
|
16.408
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr6_-_32557431
|
15.747
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr12_-_16759939
|
14.475
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_138211054
|
14.396
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr9_-_110252045
|
13.119
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_91573264
|
12.924
|
NM_133503
|
DCN
|
decorin
|
|
chr4_+_62362838
|
12.129
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr17_-_2996289
|
11.546
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr12_-_91398744
|
11.174
|
|
EPYC
|
epiphycan
|
|
chr1_+_198608216
|
9.910
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_182357757
|
9.525
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_+_122044010
|
9.268
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr8_+_1952875
|
9.263
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr3_+_35721115
|
9.146
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_-_91576579
|
9.077
|
|
DCN
|
decorin
|
|
chr12_-_16759733
|
8.655
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr15_+_32933869
|
8.270
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr6_-_32497925
|
8.121
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr14_+_29241989
|
7.866
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr12_-_16760935
|
7.831
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr20_-_1472168
|
7.754
|
NM_001122962
NM_001134836
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr6_-_24583454
|
7.644
|
NM_001252328
|
KIAA0319
|
KIAA0319
|
|
chr12_-_53601051
|
7.611
|
|
ITGB7
|
integrin, beta 7
|
|
chr7_+_80267922
|
7.347
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_-_114466397
|
7.194
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr8_+_27179918
|
6.873
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chrX_+_103031753
|
6.872
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr12_-_53600999
|
6.853
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chrX_+_103031797
|
6.780
|
|
PLP1
|
proteolipid protein 1
|
|
chr1_-_150738232
|
6.750
|
|
CTSS
|
cathepsin S
|
|
chr9_+_125132823
|
6.729
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr9_+_100263461
|
6.347
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr1_+_10509775
|
6.215
|
NM_001302
|
CORT
|
cortistatin
|
|
chr14_-_106725620
|
6.202
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr12_-_371994
|
6.099
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr2_-_211179765
|
6.036
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr11_-_114466471
|
5.961
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr6_+_131958441
|
5.930
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chrX_+_135230736
|
5.872
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_189839132
|
5.862
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_-_163113129
|
5.734
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_182357871
|
5.720
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_-_176733901
|
5.715
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_+_88720701
|
5.707
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr12_-_91576805
|
5.679
|
NM_001920
|
DCN
|
decorin
|
|
chr6_-_90024936
|
5.675
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr8_+_22210311
|
5.665
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr3_+_73110809
|
5.598
|
NM_018029
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2
|
|
chr3_-_121379701
|
5.576
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr2_+_189839078
|
5.519
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_241695433
|
5.493
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr1_-_150738255
|
5.391
|
|
CTSS
|
cathepsin S
|
|
chr1_+_152635871
|
5.371
|
NM_178430
|
LCE2D
|
late cornified envelope 2D
|
|
chr17_-_2966900
|
5.243
|
NM_014566
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5
|
|
chr4_-_88450601
|
5.109
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr14_+_92788924
|
5.033
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr2_-_178787522
|
5.032
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr5_+_147443534
|
5.028
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr12_-_16759531
|
4.899
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_152974222
|
4.868
|
NM_001097589
NM_005416
|
SPRR3
|
small proline-rich protein 3
|
|
chr12_-_10542616
|
4.826
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr17_-_7493389
|
4.793
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr8_-_110693233
|
4.759
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_174769034
|
4.754
|
NM_001243765
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr4_-_87028805
|
4.750
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_+_189839113
|
4.711
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_+_54398473
|
4.684
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr6_+_55039070
|
4.662
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr5_+_140165875
|
4.643
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr17_+_41003184
|
4.529
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr4_+_106631966
|
4.511
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr14_-_106494166
|
4.507
|
|
IGHV4-31
IGHA1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
|
|
chr6_-_133084585
|
4.498
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr12_+_18414473
|
4.469
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr12_+_101988746
|
4.467
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr17_+_56315786
|
4.438
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr9_-_93405066
|
4.399
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr7_+_80267972
|
4.393
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chrX_+_110366304
|
4.360
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr12_-_45307710
|
4.358
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr17_-_29624249
|
4.309
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr5_+_54320083
|
4.286
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr1_-_226926875
|
4.262
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr21_+_43619798
|
4.220
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr14_-_107131217
|
4.199
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr3_+_361365
|
4.175
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr4_-_100242487
|
4.145
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr10_-_21186530
|
4.121
|
NM_006393
|
NEBL
|
nebulette
|
|
chr1_-_160681552
|
4.116
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr13_-_38172862
|
4.101
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr4_+_71587674
|
4.069
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr14_-_101036130
|
4.059
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr1_-_163113938
|
4.048
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_-_91398757
|
4.041
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr17_+_74732629
|
4.041
|
NM_001242534
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr19_-_56348127
|
4.038
|
NM_145007
|
NLRP11
|
NLR family, pyrin domain containing 11
|
|
chr7_-_100076754
|
3.966
|
NM_030935
|
TSC22D4
|
TSC22 domain family, member 4
|
|
chr19_-_21950322
|
3.955
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr12_-_16759430
|
3.940
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_89829435
|
3.934
|
NM_198460
|
GBP6
|
guanylate binding protein family, member 6
|
|
chr4_-_100273831
|
3.928
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr14_-_107113943
|
3.922
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_70847817
|
3.865
|
NM_002727
|
SRGN
|
serglycin
|
|
chr5_+_140588267
|
3.811
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr6_+_28234787
|
3.744
|
NM_001023560
NM_001111039
NM_152736
|
ZNF187
|
zinc finger protein 187
|
|
chr3_+_178276487
|
3.730
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chrX_+_83116153
|
3.714
|
NM_021118
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1
|
|
chr17_-_19290471
|
3.707
|
NM_001198695
NM_002404
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr2_+_132480063
|
3.677
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr16_-_30934437
|
3.646
|
|
FBXL19-AS1
|
FBXL19 antisense RNA 1 (non-protein coding)
|
|
chr1_-_150738292
|
3.599
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr1_-_89591685
|
3.565
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr20_+_5891973
|
3.562
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr19_+_21264952
|
3.536
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr1_+_22979681
|
3.532
|
NM_000491
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr18_+_61442611
|
3.508
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr9_+_105757592
|
3.505
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr3_+_133494275
|
3.500
|
|
TF
|
transferrin
|
|
chr8_+_19796581
|
3.487
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr13_-_41556188
|
3.483
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_-_21487831
|
3.475
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr15_+_42066690
|
3.419
|
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr7_+_139528951
|
3.413
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr14_-_106725232
|
3.405
|
|
ZCWPW2
IGHV3-23
IGKV3-20
IGK@
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
immunoglobulin kappa locus
|
|
chr14_-_106733392
|
3.405
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr6_-_136847609
|
3.401
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr11_+_55029657
|
3.396
|
NM_024114
|
TRIM48
|
tripartite motif containing 48
|
|
chr11_-_83984230
|
3.395
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr22_+_51041561
|
3.376
|
NM_016431
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr11_-_13517525
|
3.366
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr21_-_31538970
|
3.360
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr3_-_9920633
|
3.356
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr12_-_52828105
|
3.348
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr2_-_152830448
|
3.328
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr1_-_59147727
|
3.313
|
|
|
|
|
chr2_+_166095911
|
3.312
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr6_-_49604524
|
3.307
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr9_-_13250313
|
3.268
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr10_+_225932
|
3.265
|
NM_001202467
NM_001202468
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr2_+_74120132
|
3.247
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr12_-_15114495
|
3.219
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr4_-_153273683
|
3.217
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr14_+_70918873
|
3.216
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr4_-_186577858
|
3.188
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_132318980
|
3.176
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr14_-_61748510
|
3.166
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr4_+_71337833
|
3.164
|
NM_152291
|
MUC7
|
mucin 7, secreted
|
|
chr14_+_39703119
|
3.160
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr2_-_2334887
|
3.135
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr6_+_27107060
|
3.126
|
NM_003495
|
HIST2H4B
HIST1H4I
|
histone cluster 2, H4b
histone cluster 1, H4i
|
|
chr1_+_209929378
|
3.092
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr20_-_18477811
|
3.086
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr3_-_42305438
|
3.061
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr5_-_160279045
|
3.027
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr1_+_196621007
|
3.025
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr11_-_84634437
|
3.012
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_+_16830786
|
3.004
|
NM_001007525
|
NWD1
|
NACHT and WD repeat domain containing 1
|
|
chr10_+_51549552
|
2.992
|
NM_002443
NM_138634
|
MSMB
|
microseminoprotein, beta-
|
|
chr2_+_169937616
|
2.991
|
NM_001142271
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr2_+_113931565
|
2.984
|
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr14_-_106746056
|
2.960
|
|
|
|
|
chr7_+_29519485
|
2.959
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr4_+_70796798
|
2.941
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chr14_-_106405633
|
2.931
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr16_+_72097124
|
2.928
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr12_+_25205488
|
2.915
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr20_-_1974393
|
2.898
|
NM_001190900
|
PDYN
|
prodynorphin
|
|
chr10_+_49609641
|
2.891
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr1_-_25747276
|
2.871
|
NM_020485
NM_138616
NM_138617
NM_138618
|
RHCE
RHD
|
Rh blood group, CcEe antigens
Rh blood group, D antigen
|
|
chr2_+_74120092
|
2.860
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr19_-_48759141
|
2.846
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr1_-_93399531
|
2.834
|
|
|
|
|
chr4_-_80329354
|
2.823
|
NM_033214
|
GK2
|
glycerol kinase 2
|
|
chr6_-_46293404
|
2.804
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_-_71012750
|
2.794
|
NM_001851
|
COL9A1
|
collagen, type IX, alpha 1
|
|
chr12_-_25150372
|
2.739
|
NM_001101339
|
C12orf77
|
chromosome 12 open reading frame 77
|
|
chr6_-_136847078
|
2.697
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr4_-_15940362
|
2.692
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr18_+_61554938
|
2.689
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr10_-_62493282
|
2.673
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_+_113931549
|
2.662
|
NM_012455
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr12_-_9760481
|
2.645
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr9_-_95298251
|
2.644
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr10_-_69597768
|
2.632
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr3_-_9291251
|
2.626
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr10_-_22498911
|
2.605
|
NM_001199938
|
EBLN1
|
endogenous Bornavirus-like nucleoprotein 1
|
|
chr15_+_66874506
|
2.605
|
|
|
|
|
chr6_+_14117486
|
2.582
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr14_-_106452693
|
2.579
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_+_169757749
|
2.563
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr17_-_66951381
|
2.539
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chrX_+_91090459
|
2.532
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr5_+_156607847
|
2.512
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr5_-_95748688
|
2.505
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr18_-_24445652
|
2.494
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr4_-_68749698
|
2.481
|
NM_004262
|
TMPRSS11D
|
transmembrane protease, serine 11D
|
|
chr8_+_27183047
|
2.409
|
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr8_-_86253863
|
2.386
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|