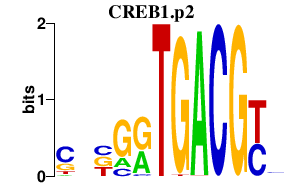
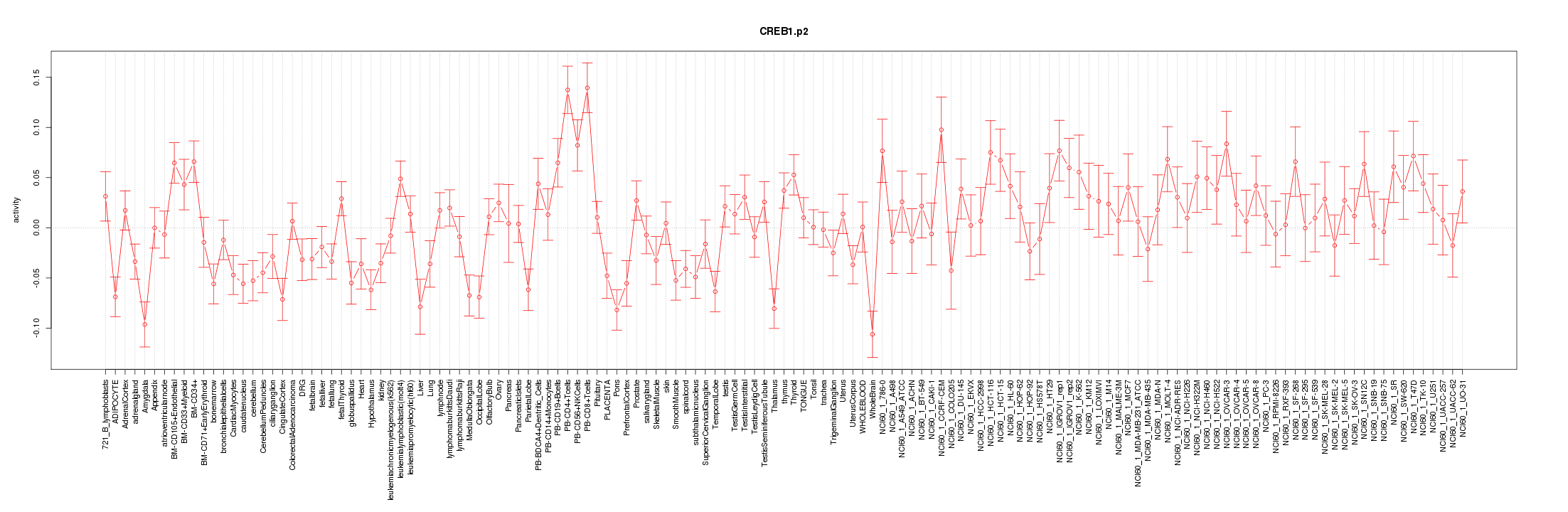
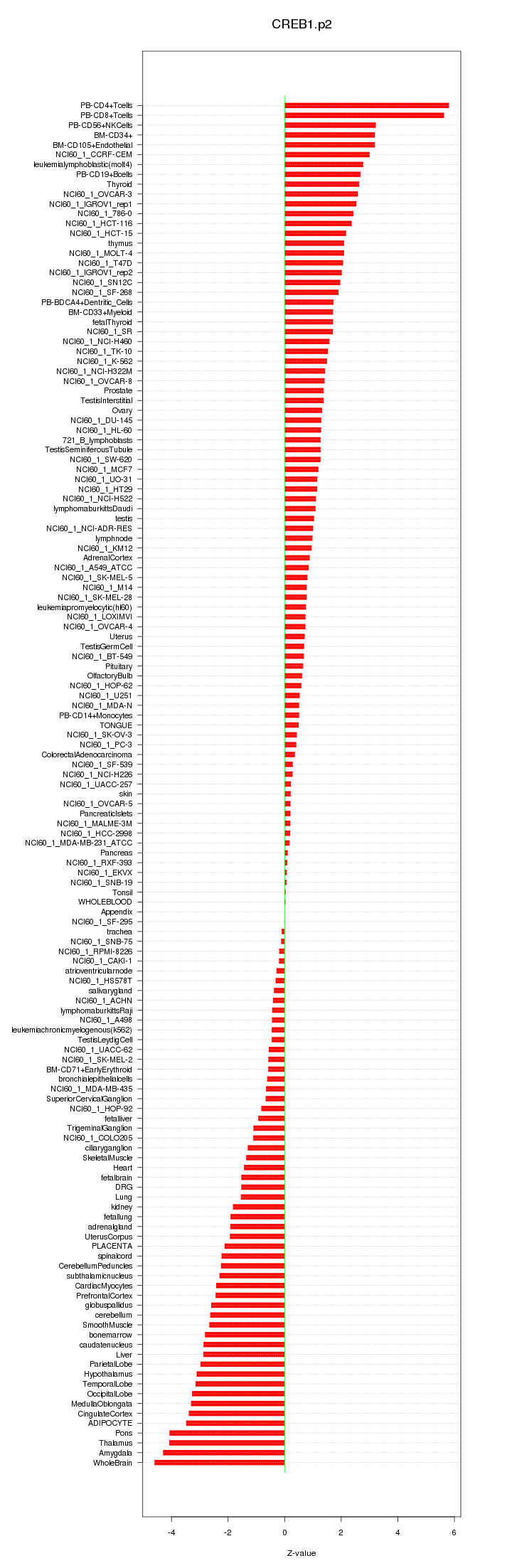
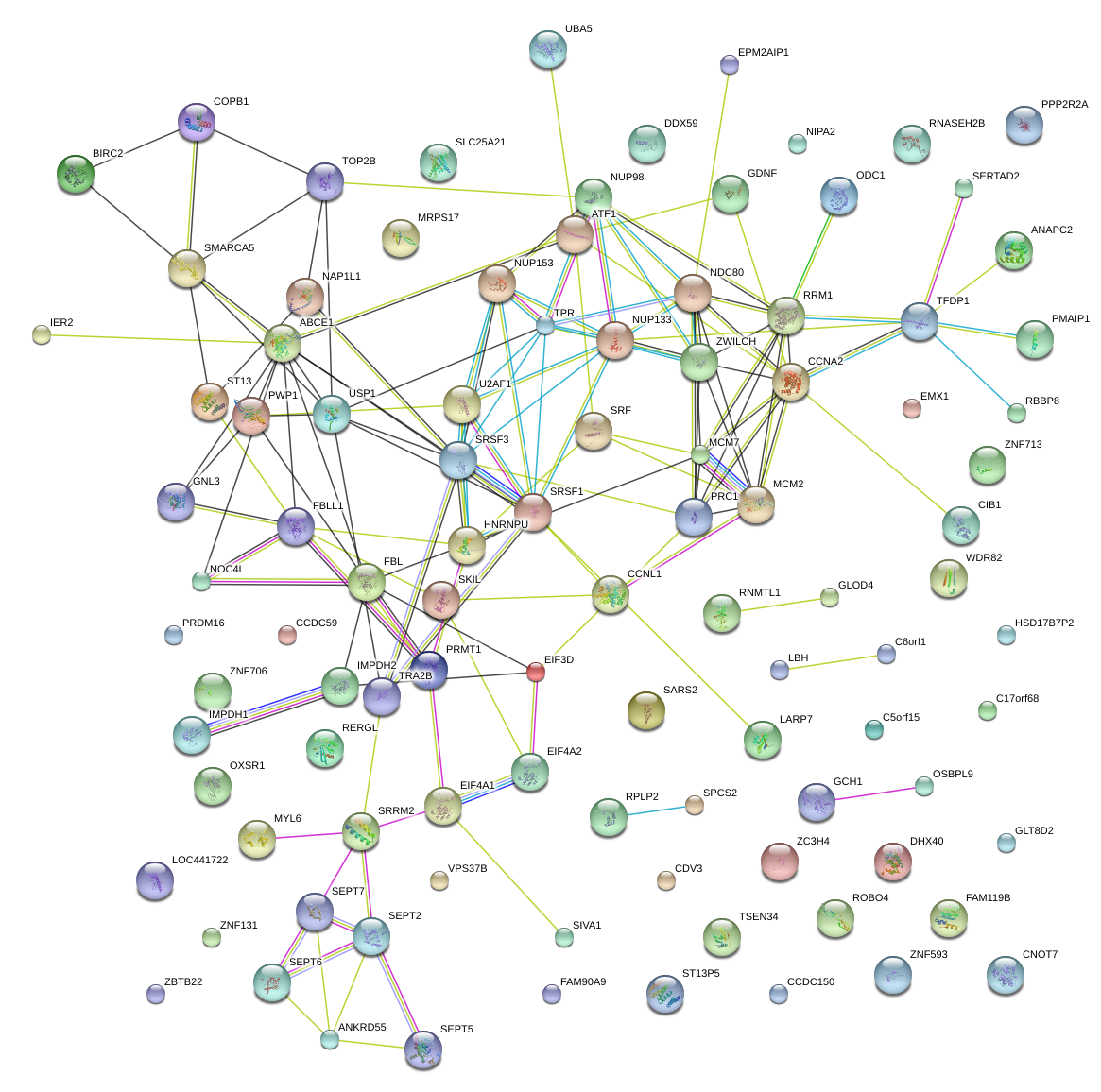
|
chr8_-_17104266
|
36.214
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr8_-_17104103
|
34.043
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr3_-_49066858
|
34.033
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr3_-_49066760
|
33.510
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr3_-_49066813
|
31.594
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr3_-_156877882
|
29.235
|
|
CCNL1
|
cyclin L1
|
|
chr12_-_123380608
|
24.120
|
|
|
|
|
chr2_-_10588273
|
23.163
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr3_+_52719935
|
22.468
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr2_-_10588451
|
22.314
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr3_+_170075521
|
19.999
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_127317271
|
19.714
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr3_+_127317274
|
19.088
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_245025958
|
17.991
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr7_+_56019552
|
17.798
|
NM_015969
|
MRPS17
|
mitochondrial ribosomal protein S17
|
|
chr17_-_685385
|
17.264
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr19_-_40336944
|
17.119
|
|
FBL
|
fibrillarin
|
|
chr19_-_40337018
|
15.990
|
NM_001436
|
FBL
|
fibrillarin
|
|
chr12_-_76478347
|
15.969
|
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr5_+_10353980
|
15.882
|
|
|
|
|
chr22_-_36925270
|
15.527
|
NM_003753
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr3_+_127317212
|
15.173
|
NM_004526
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr22_-_36925212
|
14.989
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr19_-_40336997
|
14.881
|
|
FBL
|
fibrillarin
|
|
chr19_-_47617008
|
14.223
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr17_+_57642885
|
14.130
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_+_62902735
|
13.997
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr16_+_2802683
|
13.968
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr12_+_108079516
|
13.958
|
NM_007062
|
PWP1
|
PWP1 homolog (S. cerevisiae)
|
|
chr1_-_186344801
|
13.936
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr2_-_64881040
|
13.655
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr1_+_52082755
|
13.617
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr1_+_62902653
|
13.526
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr11_+_4116059
|
13.351
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr12_+_56552141
|
13.112
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr15_-_91537724
|
13.098
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr3_+_38206968
|
13.047
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr4_+_113558119
|
12.773
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr18_+_20513277
|
12.743
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr18_+_57567191
|
12.735
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr19_+_50180492
|
12.632
|
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr22_-_41252646
|
12.606
|
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein)
|
|
chr19_+_13263890
|
12.561
|
|
IER2
|
immediate early response 2
|
|
chr16_+_2802656
|
12.521
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr13_+_51484103
|
12.484
|
|
RNASEH2B
|
ribonuclease H2, subunit B
|
|
chr3_-_52312655
|
12.481
|
NM_025222
|
WDR82
|
WD repeat domain 82
|
|
chr5_-_133304274
|
12.480
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr2_+_242255365
|
12.418
|
|
SEPT2
|
septin 2
|
|
chr11_+_4115923
|
12.304
|
NM_001033
|
RRM1
|
ribonucleotide reductase M1
|
|
chr15_+_66797632
|
12.195
|
|
ZWILCH
|
Zwilch, kinetochore associated, homolog (Drosophila)
|
|
chr17_+_685512
|
12.173
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr22_-_36925138
|
12.152
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr4_+_146019368
|
12.107
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr9_-_140083010
|
11.981
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr14_-_55369163
|
11.933
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr12_+_132629014
|
11.922
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr11_+_4116077
|
11.820
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr4_-_122744978
|
11.792
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr6_-_17706617
|
11.712
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr19_+_50180408
|
11.675
|
NM_001207042
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr16_+_2802708
|
11.617
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr12_-_123380682
|
11.584
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr12_-_82752154
|
11.529
|
NM_014167
|
CCDC59
|
coiled-coil domain containing 59
|
|
chr11_-_14521328
|
11.498
|
|
COPB1
|
coatomer protein complex, subunit beta 1
|
|
chr12_-_76478410
|
11.417
|
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr4_+_144435076
|
11.398
|
|
|
|
|
chr19_-_14529853
|
11.357
|
|
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
|
|
chr17_+_685606
|
11.311
|
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr21_-_44527603
|
11.251
|
|
U2AF1
|
U2 small nuclear RNA auxiliary factor 1
|
|
chr2_+_30454396
|
11.219
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr3_-_185655833
|
11.167
|
NM_001243879
NM_004593
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr1_+_26496382
|
11.054
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr13_+_114239047
|
11.002
|
NM_007111
|
TFDP1
|
transcription factor Dp-1
|
|
chr11_+_74660290
|
10.998
|
NM_014752
|
LOC653566
SPCS2
|
signal peptidase complex subunit 2 homolog pseudogene
signal peptidase complex subunit 2 homolog (S. cerevisiae)
|
|
chr18_+_2571587
|
10.949
|
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr16_+_2802634
|
10.941
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_-_229644077
|
10.843
|
NM_018230
|
NUP133
|
nucleoporin 133kDa
|
|
chr5_+_43121641
|
10.800
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|
|
chr4_-_122744782
|
10.790
|
|
CCNA2
|
cyclin A2
|
|
chr19_+_54694118
|
10.740
|
NM_024075
|
TSEN34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae)
|
|
chr18_+_2571509
|
10.736
|
NM_006101
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr17_-_8151373
|
10.732
|
NM_025099
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr21_-_44527681
|
10.727
|
NM_001025203
NM_001025204
NM_006758
|
U2AF1
|
U2 small nuclear RNA auxiliary factor 1
|
|
chr4_+_144434555
|
10.665
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr15_-_23034307
|
10.645
|
|
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2
|
|
chr3_-_25705719
|
10.613
|
NM_001068
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr15_-_90777144
|
10.608
|
|
CIB1
|
calcium and integrin binding 1 (calmyrin)
|
|
chr8_+_26149004
|
10.584
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr3_+_133293256
|
10.551
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr11_+_809985
|
10.535
|
|
RPLP2
|
ribosomal protein, large, P2
|
|
chr22_-_41252636
|
10.530
|
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein)
|
|
chr6_+_36562135
|
10.523
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr11_+_102217947
|
10.439
|
NM_001166
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr17_+_7476023
|
10.434
|
NM_001204510
NM_001416
|
EIF4A1
|
eukaryotic translation initiation factor 4A1
|
|
chr4_+_146019486
|
10.428
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr3_+_38207247
|
10.422
|
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr11_-_3818971
|
10.399
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chr1_-_200638996
|
10.363
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr14_+_105219466
|
10.353
|
NM_006427
NM_021709
|
SIVA1
|
SIVA1, apoptosis-inducing factor
|
|
chr12_+_51157636
|
10.342
|
NM_005171
|
ATF1
|
activating transcription factor 1
|
|
chr22_-_36925175
|
10.333
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr17_-_56084611
|
10.172
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr10_+_70715883
|
10.152
|
NM_004728
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21
|
|
chr8_+_26149043
|
10.138
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr16_-_2014826
|
9.930
|
NM_002952
|
RPS2
|
ribosomal protein S2
|
|
chr9_-_70490145
|
9.928
|
|
CBWD5
|
COBW domain containing 5
|
|
chr12_+_70636773
|
9.906
|
NM_014515
NM_001199302
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr11_-_6633400
|
9.851
|
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr11_-_67188614
|
9.833
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr1_+_171454636
|
9.783
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr7_+_107531585
|
9.757
|
NM_000108
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr4_-_103748254
|
9.648
|
NM_181892
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr19_-_9546154
|
9.633
|
NM_006631
NM_198058
|
ZNF266
|
zinc finger protein 266
|
|
chr9_-_70490106
|
9.597
|
|
CBWD5
CBWD3
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 6
|
|
chr1_+_62902708
|
9.554
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr9_-_178913
|
9.544
|
NM_001145355
NM_001145356
NM_018491
|
CBWD1
|
COBW domain containing 1
|
|
chr9_-_70490138
|
9.497
|
|
CBWD6
|
COBW domain containing 6
|
|
chr9_-_70490123
|
9.478
|
|
CBWD3
|
COBW domain containing 3
|
|
chr7_-_137686814
|
9.325
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr16_+_2802673
|
9.280
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr5_-_148930912
|
9.276
|
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr4_+_110354927
|
9.254
|
NM_001042734
NM_006323
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr16_-_67694602
|
9.235
|
NM_001082486
NM_001082487
NM_022914
|
ACD
|
adrenocortical dysplasia homolog (mouse)
|
|
chr17_+_7155371
|
9.192
|
NM_203415
NM_015362
NM_203413
NM_203414
|
C17orf81
|
chromosome 17 open reading frame 81
|
|
chr11_+_74660317
|
9.104
|
|
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae)
|
|
chr9_-_70490164
|
9.085
|
NM_001024916
NM_201453
|
CBWD5
CBWD3
|
COBW domain containing 5
COBW domain containing 3
|
|
chr21_-_40720938
|
9.072
|
|
HMGN1
|
high mobility group nucleosome binding domain 1
|
|
chr12_-_123380628
|
9.051
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr5_+_44808967
|
9.040
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr17_+_15903107
|
9.009
|
|
TTC19
|
tetratricopeptide repeat domain 19
|
|
chr19_+_36036448
|
8.995
|
NM_001242597
NM_001242598
NM_032635
|
TMEM147
|
transmembrane protein 147
|
|
chr4_+_110354825
|
8.993
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr17_-_2240677
|
8.975
|
NM_018128
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr14_-_61447711
|
8.950
|
NM_020810
|
TRMT5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae)
|
|
chr19_-_8070468
|
8.919
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr19_-_33166057
|
8.817
|
NM_032139
|
ANKRD27
|
ankyrin repeat domain 27 (VPS9 domain)
|
|
chr6_+_29691203
|
8.802
|
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr11_-_14521386
|
8.772
|
NM_016451
NM_001144062
NM_001144061
|
COPB1
|
coatomer protein complex, subunit beta 1
|
|
chr1_-_229643996
|
8.739
|
|
NUP133
|
nucleoporin 133kDa
|
|
chr8_-_131028646
|
8.738
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr11_+_18416111
|
8.729
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr6_-_109703553
|
8.695
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr7_+_107531618
|
8.678
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr21_-_44846927
|
8.669
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr8_+_26149080
|
8.632
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr1_-_35658731
|
8.626
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr14_+_50359816
|
8.626
|
|
ARF6
|
ADP-ribosylation factor 6
|
|
chr7_+_107531616
|
8.626
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr14_+_35452103
|
8.603
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr14_+_50359829
|
8.565
|
|
ARF6
|
ADP-ribosylation factor 6
|
|
chr9_-_69262517
|
8.561
|
NM_001085457
|
CBWD5
CBWD6
|
COBW domain containing 5
COBW domain containing 6
|
|
chr5_-_32174397
|
8.530
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr4_-_69215699
|
8.529
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr6_+_36562139
|
8.499
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr6_-_18264638
|
8.460
|
NM_001134709
NM_003472
|
DEK
|
DEK oncogene
|
|
chr7_-_26240364
|
8.458
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr5_-_32174385
|
8.455
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr3_+_128598471
|
8.453
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr7_-_107204336
|
8.438
|
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr9_+_70856838
|
8.425
|
NM_201453
|
CBWD5
CBWD3
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 6
|
|
chr1_+_45205533
|
8.405
|
|
KIF2C
|
kinesin family member 2C
|
|
chr6_+_47445440
|
8.379
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr8_-_131028674
|
8.369
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr11_+_18416106
|
8.357
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr22_-_47134130
|
8.353
|
NM_022766
|
CERK
|
ceramide kinase
|
|
chr11_-_6633178
|
8.351
|
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr11_+_74660321
|
8.350
|
|
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae)
|
|
chr1_-_161147209
|
8.330
|
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr19_+_59055826
|
8.305
|
NM_005762
|
TRIM28
|
tripartite motif containing 28
|
|
chr1_-_1822501
|
8.299
|
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1
|
|
chr14_-_50053068
|
8.296
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr12_+_56552044
|
8.290
|
NM_021019
NM_079423
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr2_+_114195267
|
8.275
|
NM_172003
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr22_-_29196537
|
8.226
|
NM_001079539
NM_005080
|
XBP1
|
X-box binding protein 1
|
|
chr11_+_117857062
|
8.217
|
NM_001558
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr1_+_193028523
|
8.140
|
NM_001173524
|
TROVE2
|
TROVE domain family, member 2
|
|
chr4_+_146019155
|
8.123
|
NM_001040876
NM_002940
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr20_-_5107185
|
8.117
|
NM_002592
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr11_-_6633428
|
8.043
|
NM_006284
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr14_+_35452194
|
7.998
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr13_-_31040035
|
7.976
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr1_+_202976478
|
7.958
|
|
TMEM183B
|
transmembrane protein 183B
|
|
chr5_-_32174355
|
7.903
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr16_-_46655303
|
7.896
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr16_+_2802605
|
7.853
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chrX_+_106871739
|
7.787
|
|
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1
|
|
chr4_+_110354971
|
7.786
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr7_-_26240320
|
7.774
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr17_-_36981552
|
7.769
|
NM_017748
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr11_+_34127110
|
7.745
|
NM_001144030
NM_024662
|
NAT10
|
N-acetyltransferase 10 (GCN5-related)
|
|
chr5_-_133561745
|
7.733
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr6_+_139456240
|
7.679
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr1_-_119683210
|
7.648
|
NM_015836
NM_201263
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr17_-_73775822
|
7.646
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr6_-_86352982
|
7.644
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chrX_+_106871653
|
7.628
|
NM_001204402
NM_002764
|
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1
|
|
chr1_+_45205466
|
7.624
|
NM_006845
|
KIF2C
|
kinesin family member 2C
|
|
chr16_+_2802640
|
7.585
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr7_-_26240366
|
7.579
|
NM_002137
NM_031243
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr13_-_31039966
|
7.549
|
|
HMGB1
|
high mobility group box 1
|