|
chr1_+_198608136
|
39.073
|
NM_002838
NM_080921
NM_080923
|
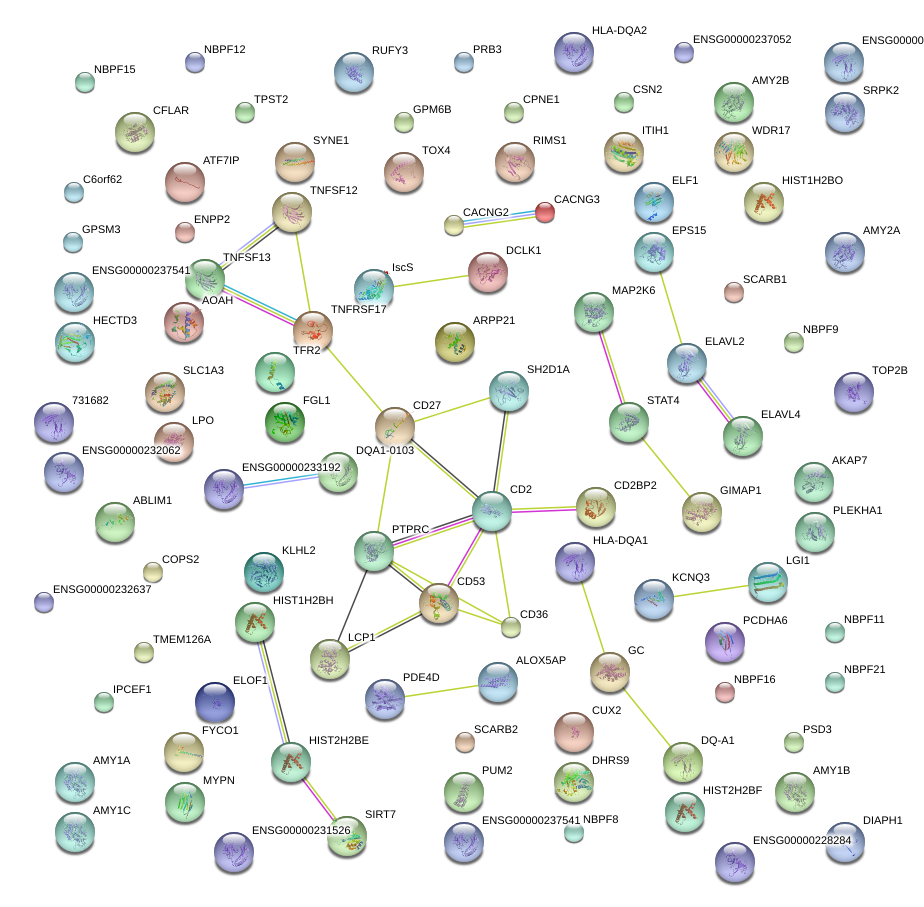
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608216
|
36.880
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608244
|
33.317
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_-_89165652
|
31.721
|
|
|
|
|
chr13_-_46756295
|
31.668
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_46756291
|
16.889
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr5_+_36606456
|
14.662
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr7_+_80231503
|
13.899
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr5_+_36606667
|
12.460
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_-_116392099
|
11.569
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr13_-_46756326
|
10.417
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr7_-_36764000
|
9.771
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr1_-_147610080
|
9.187
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr6_+_32709162
|
8.766
|
NM_020056
|
HLA-DQA1
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 1
major histocompatibility complex, class II, DQ alpha 2
|
|
chr1_-_146068244
|
8.273
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr1_+_117297056
|
7.675
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr2_+_202004997
|
7.554
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr6_-_152958533
|
7.480
|
NM_182961
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr17_+_7461591
|
7.415
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr2_-_192016293
|
7.103
|
NM_001243835
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr13_+_31309644
|
6.989
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr12_-_10562744
|
6.907
|
NM_001199805
|
KLRC4-KLRK1
|
KLRC4-KLRK1 readthrough
|
|
chr3_+_52811601
|
6.627
|
NM_002215
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr22_-_26961333
|
6.465
|
NM_001008566
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr8_-_120651019
|
6.014
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr1_-_51887740
|
5.853
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr6_-_154677868
|
5.840
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chrX_+_123480131
|
5.790
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr1_+_111413820
|
4.508
|
NM_001040033
|
CD53
|
CD53 molecule
|
|
chr12_+_6554032
|
4.253
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr8_-_18541441
|
3.966
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr13_-_36429798
|
3.832
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr2_+_169926084
|
3.811
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_+_50571963
|
3.463
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr20_-_34241830
|
3.458
|
NM_003915
|
CPNE1
|
copine I
|
|
chr12_+_74931550
|
3.372
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr4_+_71587674
|
3.245
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr7_+_80275634
|
3.236
|
NM_001127443
NM_001127444
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_+_104159953
|
3.111
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr17_-_79875995
|
3.066
|
|
SIRT7
|
sirtuin 7
|
|
chr4_-_72649734
|
3.055
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr7_-_104909442
|
2.901
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr7_-_100239136
|
2.896
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr3_+_35721115
|
2.810
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr17_-_79876027
|
2.307
|
|
SIRT7
|
sirtuin 7
|
|
chr14_+_21457964
|
2.146
|
NM_001029991
NM_022734
|
METTL17
|
methyltransferase like 17
|
|
chr13_-_41556360
|
2.103
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr17_-_79876024
|
2.080
|
|
SIRT7
|
sirtuin 7
|
|
chr3_-_25683825
|
2.051
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr10_+_69869249
|
2.048
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr4_+_176987148
|
1.838
|
|
WDR17
|
WD repeat domain 17
|
|
chr13_-_41556188
|
1.831
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr16_+_12058963
|
1.798
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chrX_-_13835313
|
1.649
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr17_+_56315786
|
1.555
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr8_-_17752847
|
1.466
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr17_+_67498391
|
1.252
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr5_-_140895407
|
1.235
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr6_+_131571298
|
1.191
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr8_-_133459508
|
1.103
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr12_-_11422640
|
1.075
|
NM_006249
|
PRB3
|
proline-rich protein BstNI subfamily 3
|
|
chr5_-_58882274
|
1.064
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_166131170
|
1.058
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr10_-_42863435
|
1.018
|
|
LOC441666
|
zinc finger protein 91 pseudogene
|
|
chr7_+_150413597
|
1.003
|
NM_001199577
NM_130759
|
GIMAP1-GIMAP5
GIMAP1
|
GIMAP1-GIMAP5 readthrough
GTPase, IMAP family member 1
|
|
chr6_-_32160637
|
0.990
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr12_+_111471827
|
0.954
|
NM_015267
|
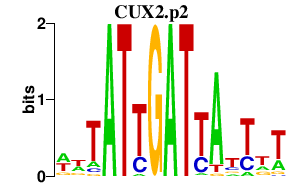
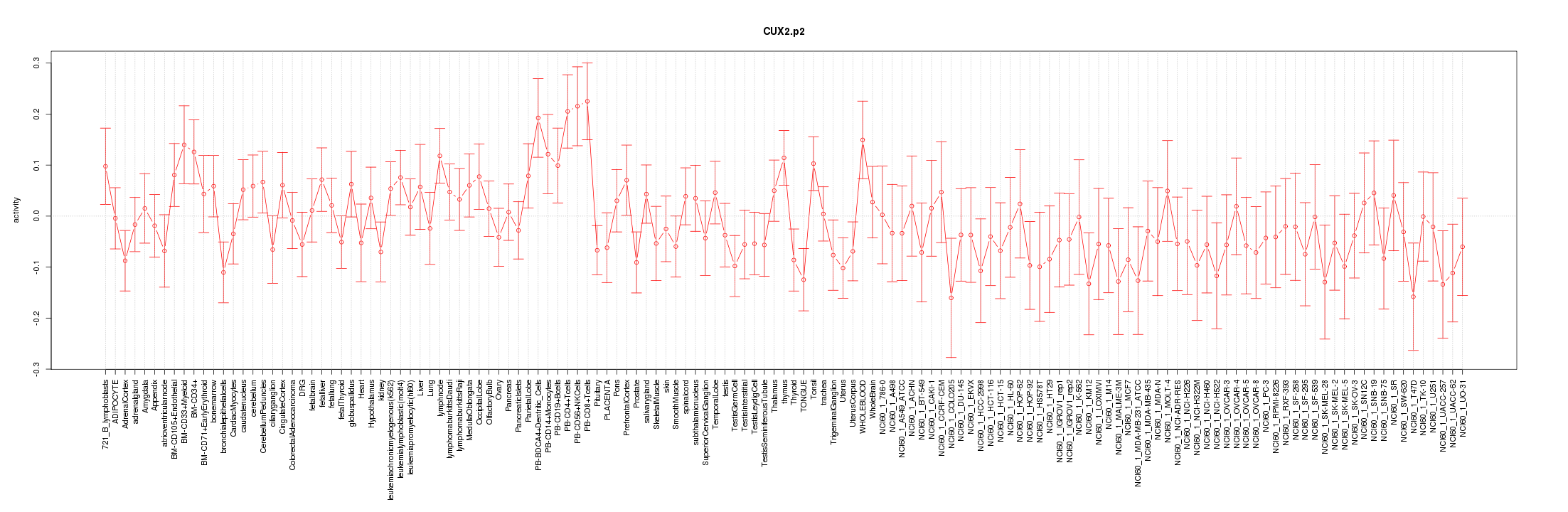
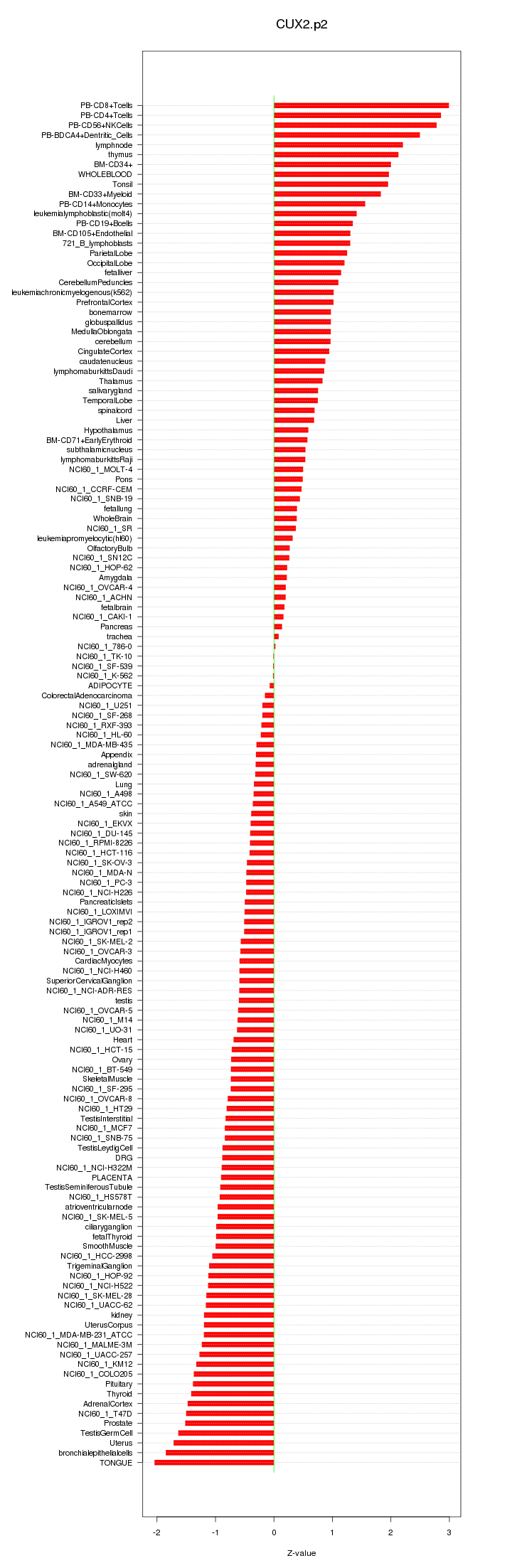
CUX2
|
cut-like homeobox 2
|
|
chr4_-_70826712
|
0.943
|
NM_001891
|
CSN2
|
casein beta
|
|
chr1_-_104238785
|
0.917
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr12_+_14538232
|
0.868
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr22_-_37098620
|
0.858
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr10_+_95517565
|
0.849
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr10_+_124151818
|
0.788
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr1_-_45477016
|
0.682
|
NM_024602
|
HECTD3
|
HECT domain containing 3
|
|
chr6_-_24719286
|
0.640
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr2_-_20512016
|
0.604
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr5_+_140247767
|
0.569
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr1_-_149399159
|
0.545
|
NM_001024599
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr6_+_72926144
|
0.528
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_134514044
|
0.354
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr8_+_18248754
|
0.320
|
NM_000015
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr17_-_67311773
|
0.313
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr11_-_30605748
|
0.290
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr8_+_17822093
|
0.286
|
|
PCM1
|
pericentriolar material 1
|
|
chr11_-_47736896
|
0.198
|
NM_024783
|
AGBL2
|
ATP/GTP binding protein-like 2
|
|
chr5_-_94224601
|
0.187
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr21_+_30400229
|
0.170
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr2_-_152830448
|
0.167
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr6_-_27799304
|
0.140
|
NM_003541
|
HIST1H4A
HIST1H4F
HIST1H4K
HIST1H4J
|
histone cluster 1, H4a
histone cluster 1, H4f
histone cluster 1, H4k
histone cluster 1, H4j
|
|
chr4_+_187187117
|
0.080
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr12_-_22089627
|
0.072
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr3_+_119500866
|
0.070
|
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr1_-_186430216
|
0.067
|
NM_002597
|
PDC
|
phosducin
|
|
chr14_-_107199126
|
0.054
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr5_-_41261539
|
0.039
|
NM_001115131
|
C6
|
complement component 6
|
|
chrX_-_106362050
|
0.039
|
NM_001171080
NM_018301
|
RBM41
|
RNA binding motif protein 41
|
|
chr18_-_31628557
|
0.024
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr3_-_170744767
|
0.013
|
NM_000340
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr18_+_59157774
|
0.002
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr2_+_132480063
|
0.001
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|