|
chr21_-_46340874
|
52.542
|
NM_000211
|
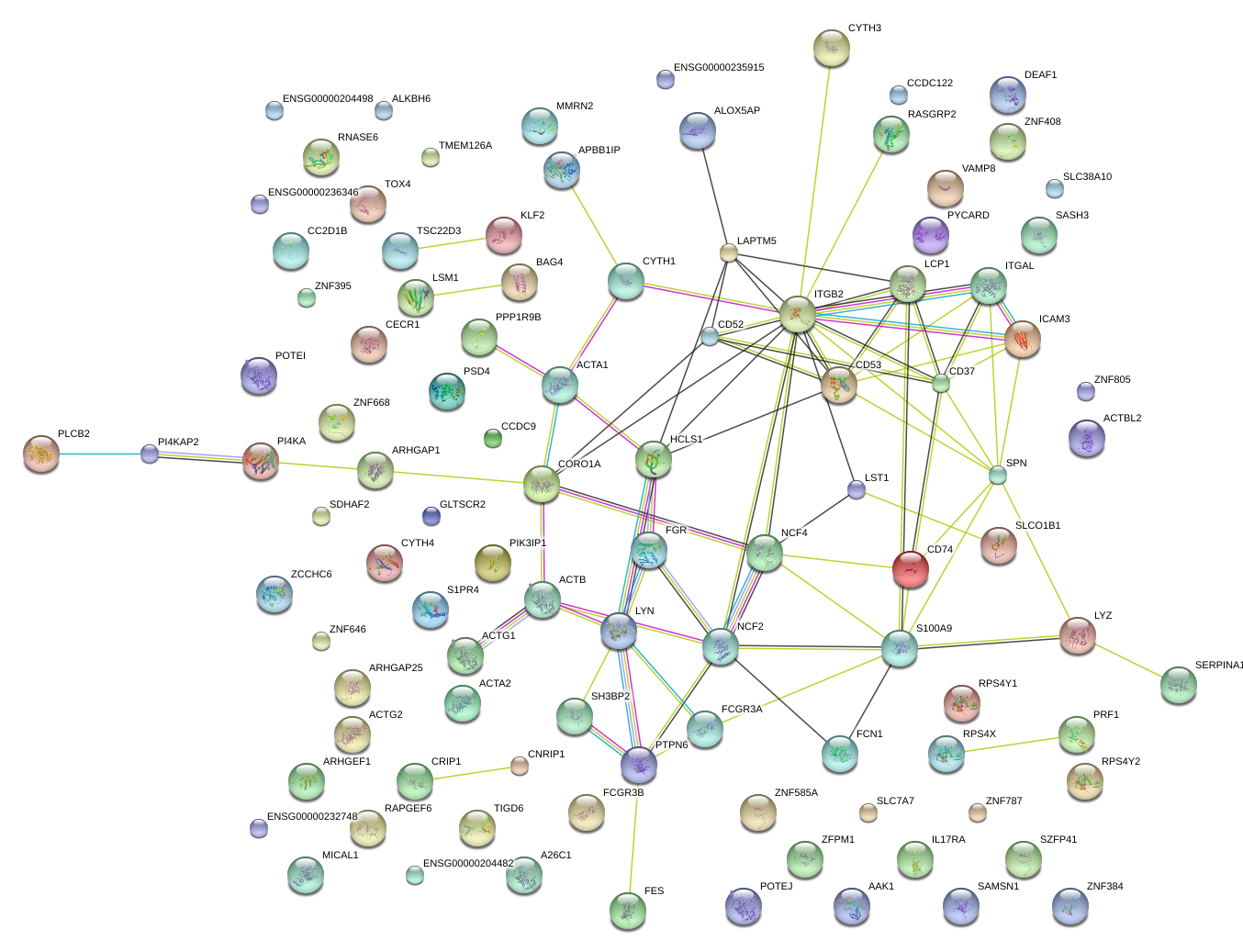
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr21_-_46340821
|
50.174
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_+_49838734
|
45.781
|
|
CD37
|
CD37 molecule
|
|
chr13_+_31287614
|
45.453
|
NM_001204406
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr19_+_49838700
|
45.450
|
|
CD37
|
CD37 molecule
|
|
chr9_-_137809710
|
42.810
|
NM_002003
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1
|
|
chr1_-_31230594
|
41.733
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr13_-_46756295
|
41.515
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr21_-_46340776
|
40.057
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr1_-_31230673
|
37.987
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr19_+_49838673
|
37.972
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr6_+_31553955
|
36.079
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr8_+_38034066
|
35.913
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr19_+_49838646
|
35.843
|
|
CD37
|
CD37 molecule
|
|
chr22_-_17680485
|
34.682
|
NM_177405
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr9_-_88969323
|
33.823
|
NM_001185059
NM_001185074
NM_024617
|
ZCCHC6
|
zinc finger, CCHC domain containing 6
|
|
chr16_+_30194925
|
33.303
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr5_-_149792276
|
32.319
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr12_-_6798419
|
31.792
|
|
ZNF384
|
zinc finger protein 384
|
|
chr19_+_42387249
|
31.566
|
NM_004706
NM_198977
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr1_+_111415721
|
31.438
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr11_+_46722564
|
31.021
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr11_-_46722139
|
30.596
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr11_-_46722160
|
30.455
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr8_-_38034168
|
29.891
|
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr14_-_94856926
|
29.886
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr19_+_16435734
|
29.363
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr1_+_111415791
|
29.085
|
|
CD53
|
CD53 molecule
|
|
chr13_-_46756326
|
28.554
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_+_47759802
|
28.298
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr19_+_47759730
|
27.621
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr11_+_46722316
|
27.494
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr19_+_42387307
|
26.457
|
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr22_+_37257004
|
26.297
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr1_-_183559669
|
25.651
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr12_+_7055736
|
24.798
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chrY_+_2709596
|
24.763
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr10_-_72362513
|
24.674
|
NM_001083116
NM_005041
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr10_-_72362441
|
24.336
|
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chrY_+_2709630
|
24.223
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr14_+_21249209
|
24.173
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr16_+_31085722
|
24.106
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr12_-_6798528
|
23.980
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr12_+_7055753
|
23.840
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr18_-_72264808
|
23.788
|
|
LOC400657
|
uncharacterized LOC400657
|
|
chr1_+_153330329
|
23.646
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr19_+_3178735
|
23.526
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chrX_-_107019001
|
23.502
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr8_+_38034296
|
23.449
|
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr1_-_161520412
|
23.174
|
NM_001127593
NM_001127595
NM_001127596
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr12_-_6798619
|
23.095
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr14_-_23284617
|
22.762
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr19_-_37701385
|
22.514
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr16_+_30483959
|
22.237
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr8_+_56792345
|
22.071
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr8_+_56792392
|
21.766
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr17_-_79269082
|
21.690
|
NM_001037984
NM_138570
|
SLC38A10
|
solute carrier family 38, member 10
|
|
chr1_-_183559463
|
21.485
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr16_+_31085751
|
21.330
|
|
ZNF646
|
zinc finger protein 646
|
|
chr16_+_30484049
|
21.313
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr5_-_149380121
|
21.239
|
NM_001243253
|
LOC728287
TIGD6
|
uncharacterized LOC728287
tigger transposable element derived 6
|
|
chr12_+_69742130
|
20.968
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr7_-_5569200
|
20.929
|
|
ACTB
|
actin, beta
|
|
chr1_-_183560010
|
20.528
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr19_-_56632644
|
20.188
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr19_-_10450262
|
20.123
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr12_+_69742140
|
19.463
|
|
LYZ
|
lysozyme
|
|
chr5_-_130970851
|
19.296
|
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr16_-_31085503
|
19.242
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr1_-_27952591
|
19.135
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr3_-_121379757
|
19.116
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr18_-_72265038
|
18.952
|
|
LOC400657
|
uncharacterized LOC400657
|
|
chr8_+_56792399
|
18.871
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_-_52831817
|
18.846
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr2_+_69001932
|
18.553
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr5_-_149792370
|
18.407
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr4_+_2813966
|
18.373
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chrX_+_128913881
|
18.179
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr22_+_37678565
|
17.744
|
|
CYTH4
|
cytohesin 4
|
|
chr16_+_29674564
|
17.694
|
|
SPN
|
sialophorin
|
|
chr14_+_105953531
|
17.690
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chrX_+_128913897
|
17.611
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr11_+_61197574
|
17.486
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr15_-_40599872
|
17.429
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr22_-_31688329
|
17.414
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr16_-_31214054
|
17.373
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chr19_+_39899749
|
17.337
|
|
|
|
|
chr3_-_121379701
|
17.079
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr10_-_88717319
|
16.929
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr1_+_26644458
|
16.570
|
|
CD52
|
CD52 molecule
|
|
chr2_-_69870837
|
16.460
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr1_-_161519817
|
16.417
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr1_-_161601752
|
16.214
|
NM_000570
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr19_-_36505102
|
16.090
|
|
ALKBH6
|
alkB, alkylation repair homolog 6 (E. coli)
|
|
chr15_+_91427664
|
16.041
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr19_+_57752051
|
15.883
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr13_-_46756291
|
15.881
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr15_-_40600118
|
15.308
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr21_-_15918635
|
15.236
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr6_-_109776942
|
15.185
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chrX_-_107018889
|
15.177
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_-_64512925
|
15.159
|
NM_153819
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr2_+_85804613
|
15.087
|
NM_003761
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chr10_+_26727692
|
15.059
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr13_-_44453825
|
15.012
|
NM_144974
|
CCDC122
|
coiled-coil domain containing 122
|
|
chr2_+_113931549
|
14.986
|
NM_012455
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr22_-_21213069
|
14.970
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr19_+_48248798
|
14.947
|
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr22_+_17565844
|
14.946
|
NM_014339
|
IL17RA
|
interleukin 17 receptor A
|
|
chr1_-_1590403
|
14.938
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr22_+_37678423
|
14.922
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chrX_-_47489703
|
14.814
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr14_-_24711767
|
14.779
|
NM_001099274
NM_012461
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr6_-_109776946
|
14.766
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr5_-_130970907
|
14.680
|
NM_001164386
NM_001164387
NM_001164388
NM_001164389
NM_001164390
NM_016340
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr19_+_48248792
|
14.488
|
NM_015710
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr19_-_36399120
|
14.422
|
|
TYROBP
|
TYRO protein tyrosine kinase binding protein
|
|
chr6_-_33267128
|
14.401
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr18_+_11851388
|
14.148
|
NM_020412
|
CHMP1B
|
charged multivesicular body protein 1B
|
|
chr6_+_31554975
|
14.107
|
NM_001166538
NM_007161
NM_205840
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_+_113931565
|
14.057
|
|
PSD4
|
pleckstrin and Sec7 domain containing 4
|
|
chr2_-_69870705
|
14.046
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr21_-_44299605
|
13.986
|
|
WDR4
|
WD repeat domain 4
|
|
chr12_-_6233684
|
13.953
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr1_+_150266261
|
13.929
|
NM_018997
NM_031901
|
MRPS21
|
mitochondrial ribosomal protein S21
|
|
chr22_-_37545944
|
13.894
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr10_+_26727651
|
13.887
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr13_+_31191829
|
13.869
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr14_-_24036532
|
13.848
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr12_-_51717835
|
13.768
|
|
BIN2
|
bridging integrator 2
|
|
chr9_+_77703412
|
13.721
|
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr19_+_16999871
|
13.661
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr7_-_102105161
|
13.616
|
|
ALKBH4
|
alkB, alkylation repair homolog 4 (E. coli)
|
|
chr6_-_133084585
|
13.525
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr12_+_54892567
|
13.518
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr3_+_47324329
|
13.501
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr15_+_81589240
|
13.452
|
NM_004513
|
IL16
|
interleukin 16
|
|
chr12_+_7060433
|
13.376
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr2_+_68961912
|
13.372
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr14_+_69865380
|
13.328
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr15_-_40600025
|
13.276
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr7_+_108210188
|
13.197
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr2_+_103035148
|
13.190
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr7_+_108210291
|
13.185
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr20_-_23066828
|
13.150
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr14_-_23284565
|
13.135
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr7_+_150264457
|
13.116
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr11_-_67771527
|
13.095
|
NM_030930
|
UNC93B1
|
unc-93 homolog B1 (C. elegans)
|
|
chr6_-_109777171
|
13.048
|
NM_001159291
NM_022765
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr14_+_69865095
|
12.984
|
NM_001252151
NM_001252152
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr12_-_51717905
|
12.874
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr7_+_150434435
|
12.742
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_+_31554475
|
12.730
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr16_+_29465821
|
12.641
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 readthrough
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr20_+_42839599
|
12.640
|
|
LOC100505783
|
uncharacterized LOC100505783
|
|
chr12_-_15114495
|
12.609
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr2_-_96811138
|
12.609
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr2_+_17935124
|
12.593
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr1_+_24882565
|
12.542
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr22_-_37640287
|
12.536
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr19_+_16296211
|
12.529
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr11_+_5710816
|
12.507
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr2_+_17935379
|
12.475
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chrX_+_48542159
|
12.461
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr14_+_75745480
|
12.396
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr6_-_32095993
|
12.389
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chrX_+_67913490
|
12.323
|
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr2_+_143886898
|
12.290
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chrX_+_67913479
|
12.286
|
NM_001142504
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr22_+_37678519
|
12.265
|
|
CYTH4
|
cytohesin 4
|
|
chr1_+_10003485
|
12.221
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr15_+_75074417
|
12.141
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr1_+_28995214
|
12.137
|
NM_006582
NM_024482
|
GMEB1
|
glucocorticoid modulatory element binding protein 1
|
|
chr19_-_5680910
|
12.132
|
NM_205767
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr19_-_13213973
|
12.131
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr16_+_69166538
|
12.120
|
|
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin)
|
|
chr12_+_6898705
|
12.096
|
|
CD4
|
CD4 molecule
|
|
chr12_+_51318515
|
12.049
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr5_+_156693051
|
12.042
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr6_-_170102098
|
11.921
|
NM_001202550
NM_182552
|
WDR27
|
WD repeat domain 27
|
|
chr1_+_158801105
|
11.888
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr16_-_30366681
|
11.803
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr16_+_28505920
|
11.797
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr5_+_118604417
|
11.793
|
NM_001077654
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr11_+_117857123
|
11.781
|
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr11_+_65190272
|
11.772
|
|
|
|
|
chr12_-_51717941
|
11.735
|
|
BIN2
|
bridging integrator 2
|
|
chr22_-_21213099
|
11.721
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr6_-_32497925
|
11.650
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr6_+_110012423
|
11.560
|
NM_014845
|
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae)
|
|
chr12_+_112856701
|
11.490
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr2_+_143886969
|
11.477
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr22_+_22786283
|
11.472
|
|
|
|
|
chr1_+_26644380
|
11.374
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr19_+_58694444
|
11.372
|
|
ZNF274
|
zinc finger protein 274
|
|
chr6_-_32095974
|
11.354
|
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr19_+_56186426
|
11.342
|
NM_001130072
NM_013333
|
EPN1
|
epsin 1
|
|
chr1_-_161519526
|
11.331
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr15_-_64648359
|
11.325
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr12_-_92539614
|
11.235
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|