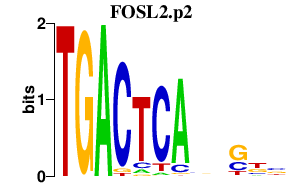
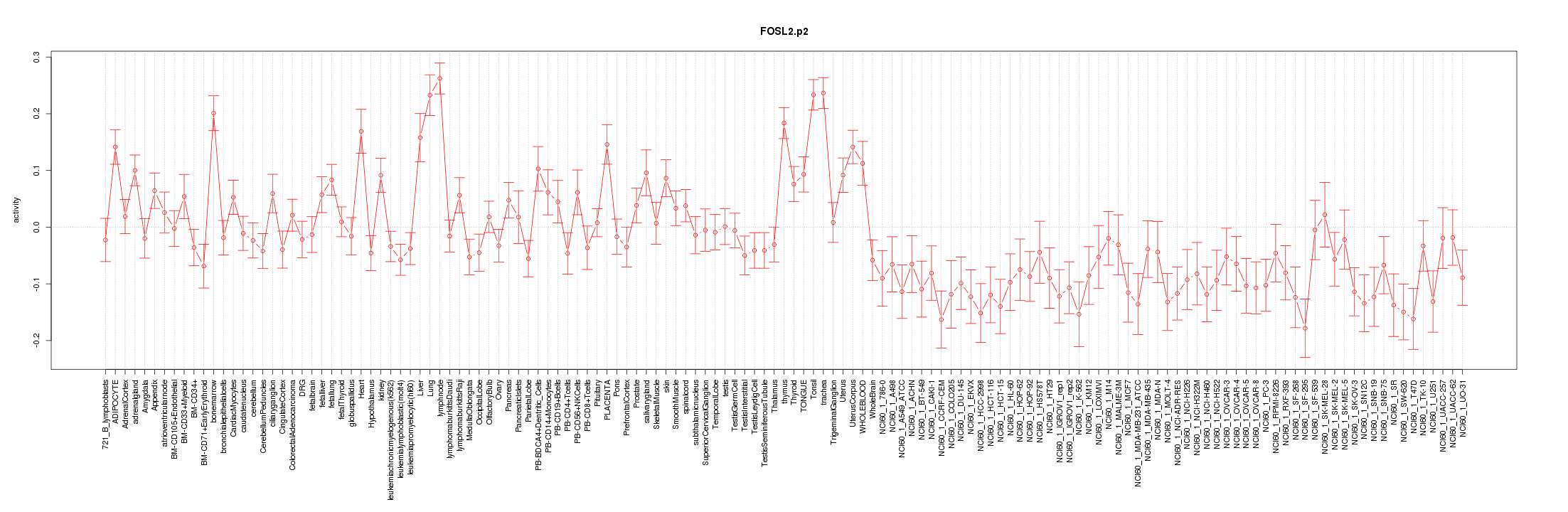
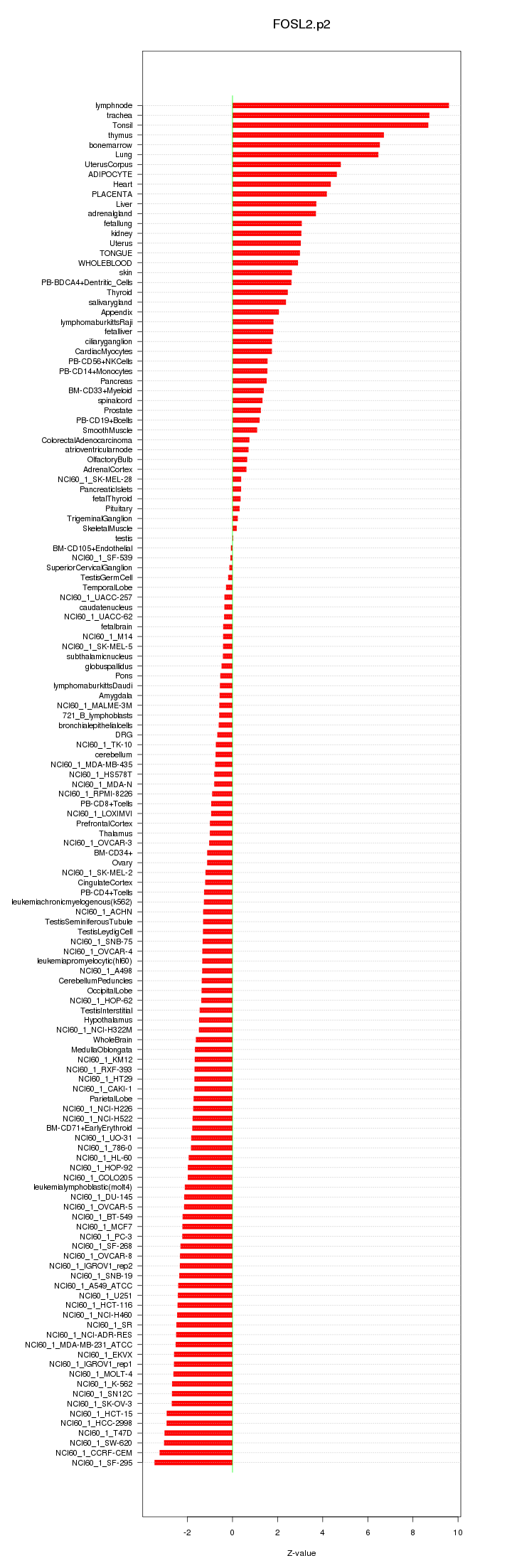
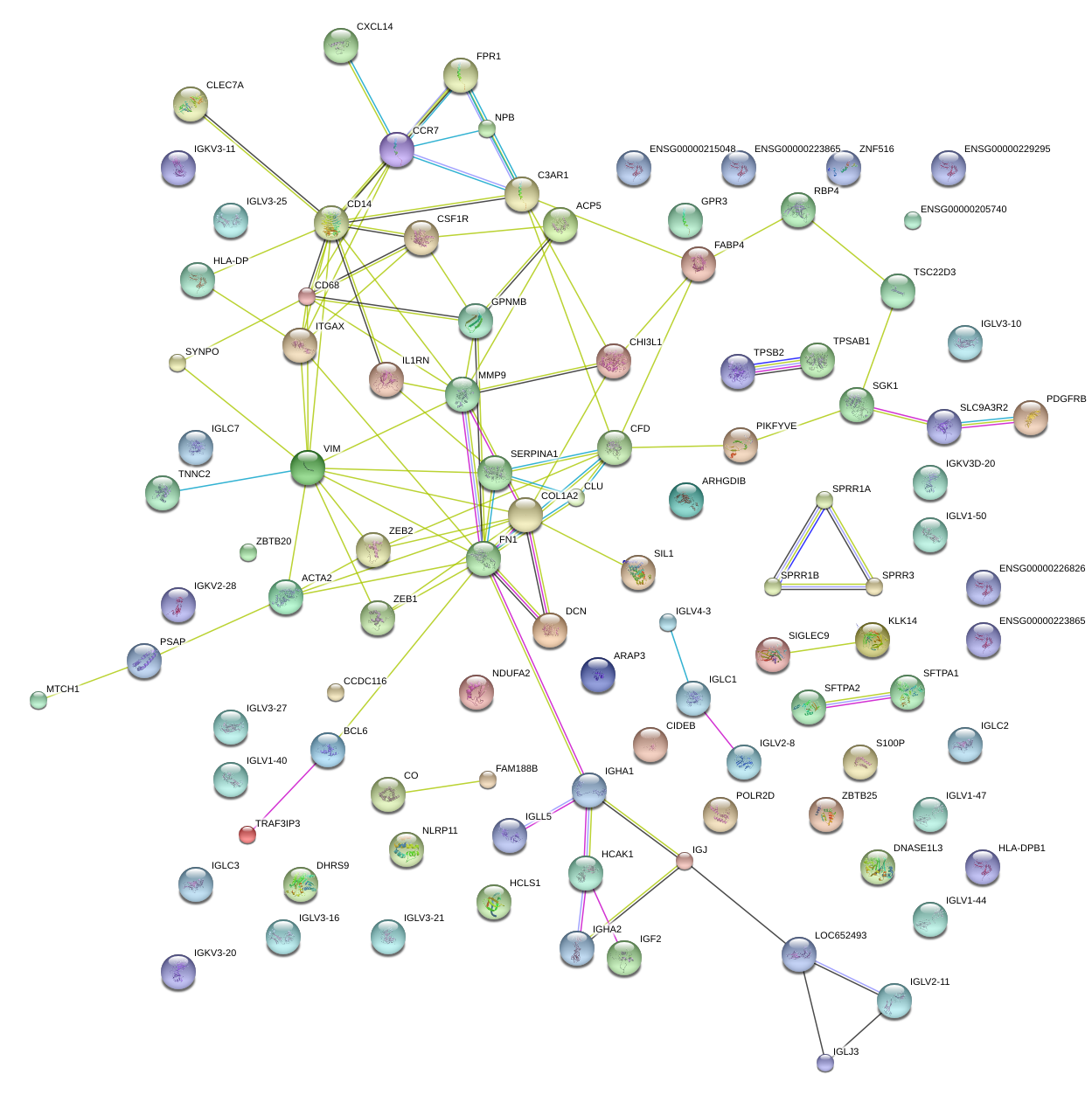
|
chr22_+_22758538
|
91.407
|
|
|
|
|
chr22_+_22735134
|
90.076
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr22_+_23040394
|
83.849
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr2_-_89292396
|
72.515
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr22_+_22569155
|
65.977
|
|
|
|
|
chr2_-_89442551
|
65.191
|
|
IGKC
IGK@
|
immunoglobulin kappa constant
immunoglobulin kappa locus
|
|
chr22_+_22735207
|
60.774
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr2_-_89513056
|
55.308
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr7_+_30960914
|
52.900
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr7_+_23286386
|
52.672
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286277
|
52.654
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286384
|
51.291
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr22_+_22723972
|
50.903
|
|
|
|
|
chr7_+_23286390
|
50.776
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr6_+_33043702
|
50.695
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr22_+_23165273
|
48.614
|
|
|
|
|
chr2_-_89619820
|
45.614
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr7_+_23286420
|
40.370
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_-_15114495
|
37.832
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr22_+_22735202
|
34.929
|
|
|
|
|
chr22_+_22712091
|
32.477
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr5_+_150020201
|
29.972
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr22_+_23229959
|
29.738
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr22_+_21986968
|
29.187
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr3_-_121379757
|
28.489
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_-_121379701
|
27.939
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr7_+_94024201
|
27.931
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr22_+_23213671
|
26.932
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr7_+_94023872
|
26.625
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr5_-_140012730
|
26.336
|
|
CD14
|
CD14 molecule
|
|
chr14_-_107179043
|
26.212
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr5_-_149535420
|
25.718
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_91573264
|
25.038
|
NM_133503
|
DCN
|
decorin
|
|
chr22_+_23063107
|
24.070
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr20_-_44455932
|
24.057
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr14_-_94855120
|
22.959
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr19_-_11688441
|
22.647
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_-_94854910
|
22.554
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr22_+_23029175
|
22.292
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr22_+_22676814
|
22.180
|
|
|
|
|
chr22_+_23010757
|
21.441
|
|
|
|
|
chr2_+_90077732
|
21.363
|
|
|
|
|
chr22_+_23154240
|
21.186
|
|
|
|
|
chr10_-_73611041
|
20.891
|
|
PSAP
|
prosaposin
|
|
chr5_-_149492934
|
20.787
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr10_-_73611005
|
20.601
|
|
PSAP
|
prosaposin
|
|
chr19_-_11688484
|
20.344
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr4_-_71532341
|
20.225
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr5_-_140013034
|
19.831
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr2_-_216259197
|
19.746
|
|
FN1
|
fibronectin 1
|
|
chr8_-_82395401
|
19.136
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr17_-_38721701
|
19.096
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr12_-_10282680
|
18.846
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr14_-_94849552
|
18.716
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr5_-_149466173
|
18.540
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr10_-_73611050
|
18.342
|
NM_001042465
NM_001042466
NM_002778
|
PSAP
|
prosaposin
|
|
chr16_+_31366519
|
18.228
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr10_-_73610958
|
17.945
|
|
PSAP
|
prosaposin
|
|
chr10_+_17270257
|
17.916
|
NM_003380
|
VIM
|
vimentin
|
|
chr8_-_27472188
|
17.850
|
|
CLU
|
clusterin
|
|
chr5_-_149466126
|
17.576
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr16_+_31366468
|
17.573
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr11_-_2162340
|
17.466
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_+_31366529
|
17.288
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr20_+_44637539
|
17.185
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr19_-_56348127
|
16.582
|
NM_145007
|
NLRP11
|
NLR family, pyrin domain containing 11
|
|
chr5_-_134914968
|
16.501
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chrX_-_106960268
|
16.363
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_-_11688508
|
16.336
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr2_+_90211704
|
15.980
|
|
|
|
|
chr14_-_24777184
|
15.963
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr4_-_71532206
|
15.790
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr6_-_134496833
|
15.676
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_-_141061798
|
15.635
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr18_-_74207145
|
15.574
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr5_-_149466094
|
15.399
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr10_-_95360959
|
15.324
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr16_-_1280162
|
15.180
|
NM_024164
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr14_-_24777160
|
14.989
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr14_-_24777153
|
14.979
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr16_+_2083264
|
14.885
|
NM_001252073
NM_001252075
NM_001252076
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr1_+_152974222
|
14.720
|
NM_001097589
NM_005416
|
SPRR3
|
small proline-rich protein 3
|
|
chr5_-_141061749
|
14.452
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr4_+_6695565
|
14.325
|
NM_005980
|
S100P
|
S100 calcium binding protein P
|
|
chr2_+_89185055
|
14.198
|
|
|
|
|
chr17_+_7482804
|
14.166
|
NM_001251
NM_001040059
|
CD68
|
CD68 molecule
|
|
chr5_-_140013285
|
14.156
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr8_-_27472298
|
14.144
|
NM_001831
|
CLU
|
clusterin
|
|
chr19_-_51587466
|
13.914
|
NM_022046
|
KLK14
|
kallikrein-related peptidase 14
|
|
chr1_+_209929378
|
13.778
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr10_+_17270469
|
13.673
|
|
VIM
|
vimentin
|
|
chr2_-_145274916
|
13.256
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_+_7482938
|
13.208
|
|
CD68
|
CD68 molecule
|
|
chr2_+_169926084
|
13.137
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_+_152956563
|
13.020
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr2_+_113875469
|
12.994
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr12_-_8219001
|
12.651
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr3_-_58200397
|
12.584
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr1_+_27719151
|
12.228
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr3_-_187454247
|
12.195
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_-_8218930
|
12.178
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr14_-_64970319
|
11.922
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr19_-_52255099
|
11.909
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr5_-_138533982
|
11.849
|
|
SIL1
|
SIL1 homolog, endoplasmic reticulum chaperone (S. cerevisiae)
|
|
chr1_-_203155867
|
11.701
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr1_-_203155910
|
11.423
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr19_+_51628136
|
11.349
|
NM_001198558
NM_014441
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9
|
|
chr19_+_859658
|
11.347
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr3_-_114343791
|
11.213
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr22_-_50968137
|
11.187
|
NM_001113756
|
TYMP
|
thymidine phosphorylase
|
|
chr17_+_7482980
|
11.186
|
|
CD68
|
CD68 molecule
|
|
chr1_-_203155827
|
11.135
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr7_+_142495142
|
11.125
|
|
|
|
|
chr6_+_36410800
|
11.082
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr16_+_1306272
|
11.001
|
NM_012217
|
TPSD1
|
tryptase delta 1
|
|
chr14_+_69726868
|
10.965
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr22_+_23134980
|
10.927
|
|
|
|
|
chr19_+_859673
|
10.730
|
|
CFD
|
complement factor D (adipsin)
|
|
chr12_-_8043743
|
10.554
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr12_-_7656285
|
10.520
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr5_-_138533998
|
10.501
|
|
SIL1
|
SIL1 homolog, endoplasmic reticulum chaperone (S. cerevisiae)
|
|
chr12_-_48152860
|
10.417
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_150480584
|
10.387
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr2_-_152830448
|
10.295
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_+_35856990
|
10.292
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr6_+_138188352
|
10.246
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_-_153348057
|
10.220
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr12_-_10021932
|
10.216
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr6_-_24877453
|
10.118
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr19_+_41903718
|
10.112
|
|
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide
|
|
chr7_+_150264457
|
10.011
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr20_-_44539542
|
9.993
|
|
PLTP
|
phospholipid transfer protein
|
|
chr19_+_50432399
|
9.933
|
NM_001193646
|
ATF5
|
activating transcription factor 5
|
|
chr16_-_1832576
|
9.871
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr7_-_98467615
|
9.747
|
|
TMEM130
|
transmembrane protein 130
|
|
chr19_+_50016606
|
9.668
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr3_+_183903970
|
9.482
|
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr5_-_138534026
|
9.425
|
NM_001037633
NM_022464
|
SIL1
|
SIL1 homolog, endoplasmic reticulum chaperone (S. cerevisiae)
|
|
chr4_-_39033981
|
9.372
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr19_-_51537996
|
9.311
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr11_+_60739114
|
9.260
|
NM_006725
|
CD6
|
CD6 molecule
|
|
chr1_-_11866079
|
9.256
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr14_-_24777064
|
9.243
|
|
|
|
|
chr12_+_10365415
|
9.179
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr1_+_17634689
|
9.163
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr17_-_34207308
|
9.011
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr1_-_182357871
|
8.983
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr19_-_49140569
|
8.934
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr1_+_154377847
|
8.917
|
|
IL6R
|
interleukin 6 receptor
|
|
chr6_+_36410543
|
8.907
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr12_+_21679190
|
8.882
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr16_-_1832571
|
8.876
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr9_+_4985082
|
8.862
|
|
JAK2
|
Janus kinase 2
|
|
chr3_-_69171536
|
8.853
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr8_+_22210311
|
8.834
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr12_-_48152180
|
8.775
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr12_+_52445185
|
8.700
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr22_+_22764094
|
8.658
|
|
|
|
|
chr3_+_183903862
|
8.608
|
NM_018358
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chrX_-_154563735
|
8.594
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr16_+_56965747
|
8.549
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr6_+_106546736
|
8.531
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_50786438
|
8.506
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr1_-_57045214
|
8.495
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_56966025
|
8.466
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr12_+_103981050
|
8.461
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr1_-_160232269
|
8.455
|
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr1_-_120311466
|
8.383
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr16_+_28996369
|
8.354
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr22_+_37309618
|
8.351
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr1_-_203155745
|
8.340
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr4_+_154178525
|
8.329
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr15_+_41136622
|
8.210
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr3_-_194090459
|
8.168
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chr2_-_64371444
|
8.127
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr9_+_132251323
|
8.090
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr2_-_85895863
|
8.050
|
NM_000542
NM_198843
|
SFTPB
|
surfactant protein B
|
|
chr8_+_11561710
|
8.043
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr15_+_89182038
|
8.015
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr16_+_30968614
|
7.991
|
NM_014712
|
SETD1A
|
SET domain containing 1A
|
|
chr1_+_150122415
|
7.968
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr19_-_19738973
|
7.899
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr1_+_150480486
|
7.858
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr2_+_68961912
|
7.849
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr1_+_150122246
|
7.846
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr6_+_154407641
|
7.829
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr9_+_132251403
|
7.791
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr19_+_21264952
|
7.779
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr12_+_7060433
|
7.752
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr17_+_42264582
|
7.705
|
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr3_+_148447966
|
7.699
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr15_+_40331417
|
7.666
|
|
LOC100131089
|
uncharacterized LOC100131089
|
|
chr1_+_154378044
|
7.632
|
|
IL6R
|
interleukin 6 receptor
|
|
chr11_-_120008801
|
7.606
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr3_+_58223228
|
7.573
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr16_+_28505920
|
7.535
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr11_+_60197061
|
7.527
|
NM_023945
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5
|
|
chr21_+_43619798
|
7.506
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr11_-_2162167
|
7.497
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr6_-_44233499
|
7.492
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|