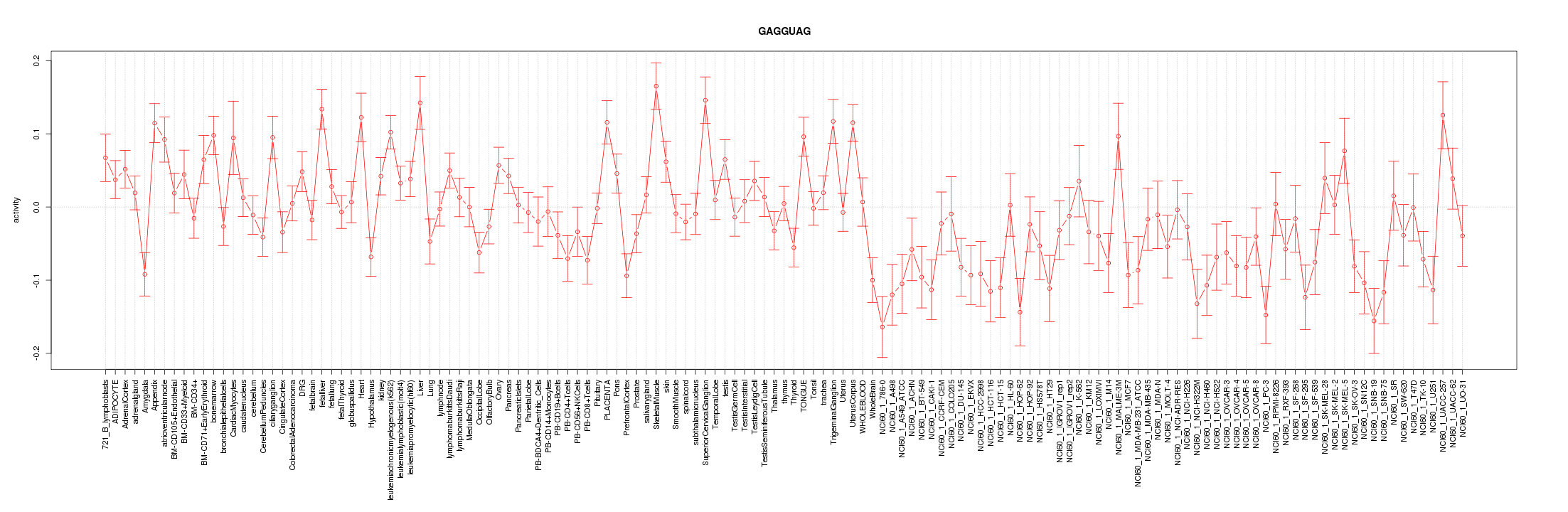
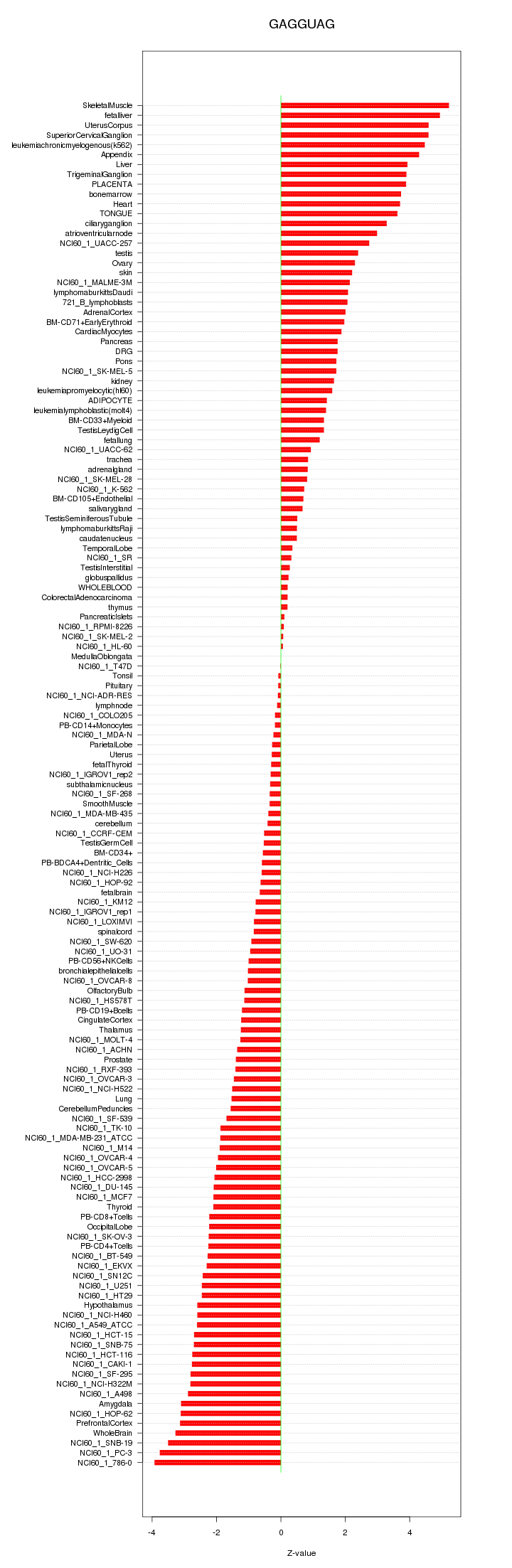
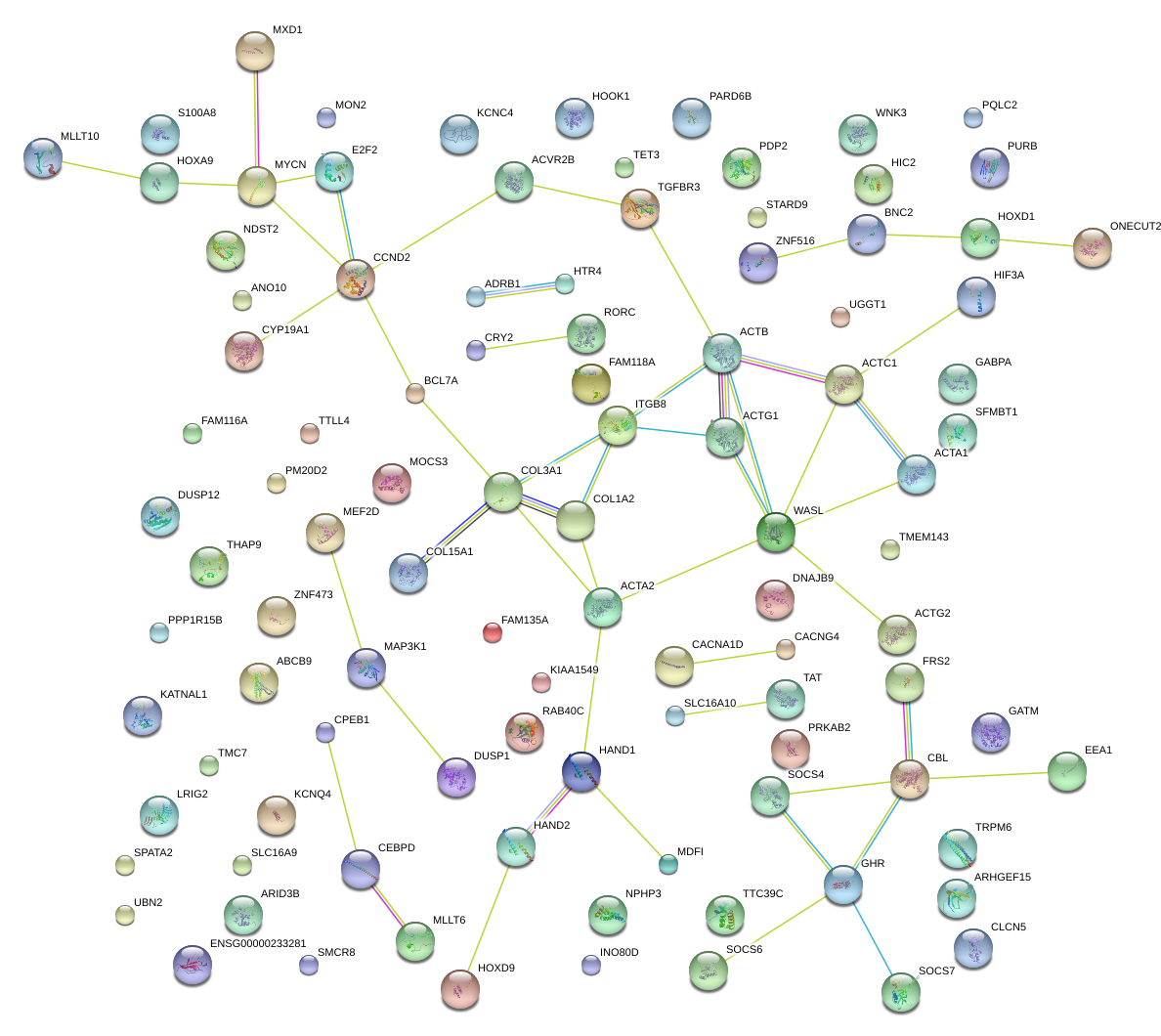
|
chr18_+_21594349
|
35.657
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr1_-_204380903
|
33.616
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr13_-_30881141
|
27.902
|
NM_001014380
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr10_-_61469322
|
27.631
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr18_+_21572736
|
27.294
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr13_-_30881556
|
27.245
|
NM_032116
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr6_+_89855745
|
19.358
|
NM_001010853
|
PM20D2
|
peptidase M20 domain containing 2
|
|
chrX_-_54384436
|
17.560
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr7_+_94023872
|
15.106
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr15_+_42867708
|
14.826
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr9_+_101705710
|
14.578
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr1_-_92371558
|
14.351
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr4_-_174451356
|
14.164
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr1_-_92351613
|
14.049
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_153363544
|
13.789
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr15_+_74833517
|
13.740
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr18_+_55102777
|
13.683
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr17_+_36507708
|
13.614
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr2_+_189839078
|
13.293
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_70142152
|
12.690
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr12_-_93323088
|
12.579
|
NM_003566
|
EEA1
|
early endosome antigen 1
|
|
chr1_-_156470506
|
12.381
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr7_-_44924946
|
12.152
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr18_-_74207145
|
12.131
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr17_+_18218569
|
11.622
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr12_+_62860531
|
11.509
|
NM_015026
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr12_+_69864065
|
11.463
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr15_-_83316621
|
11.330
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr2_+_74273449
|
11.297
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr5_-_172198160
|
11.200
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr5_-_153857820
|
11.190
|
NM_004821
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr19_+_46800296
|
11.138
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr15_-_45670955
|
11.089
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr21_+_27107257
|
10.718
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr7_+_138916230
|
10.271
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr7_-_138665853
|
10.258
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr21_+_27107322
|
10.254
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr12_+_4382882
|
10.218
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr15_-_51630735
|
10.188
|
NM_000103
NM_031226
|
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1
|
|
chr1_-_229569839
|
10.167
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr1_+_19638739
|
10.046
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr7_+_108210188
|
9.899
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr3_+_53529030
|
9.782
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr12_-_123451031
|
9.762
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr16_-_71610948
|
9.726
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr9_-_77502262
|
9.713
|
NM_001177311
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr2_+_16080639
|
9.659
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr3_-_57678594
|
9.654
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr1_+_41249479
|
9.637
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr9_-_16870719
|
9.554
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr10_-_75571521
|
9.537
|
NM_003635
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2
|
|
chr22_+_21771661
|
9.500
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr6_+_71123105
|
9.473
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr11_+_119076974
|
9.316
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr20_+_49575347
|
9.315
|
NM_014484
|
MOCS3
|
molybdenum cofactor synthesis 3
|
|
chr19_-_48867149
|
9.314
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chrX_+_49687212
|
9.307
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr6_+_111408780
|
9.222
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr1_+_110753313
|
9.180
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr3_+_38495778
|
9.168
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr7_-_123388711
|
9.124
|
NM_003941
|
WASL
|
Wiskott-Aldrich syndrome-like
|
|
chr16_+_639668
|
9.050
|
NM_001172664
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr4_+_83821835
|
9.013
|
NM_024672
|
THAP9
|
THAP domain containing 9
|
|
chr6_+_41606193
|
9.012
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr10_+_115803805
|
8.994
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr2_+_177053306
|
8.955
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr17_+_64960754
|
8.927
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr3_-_53080038
|
8.897
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr5_+_42425093
|
8.831
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr3_-_132441302
|
8.801
|
NM_153240
|
NPHP3-ACAD11
NPHP3
|
NPHP3-ACAD11 readthrough
nephronophthisis 3 (adolescent)
|
|
chr1_-_151798552
|
8.762
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr16_+_66914380
|
8.741
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr5_+_42424553
|
8.727
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr20_+_49348050
|
8.652
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr10_+_21823573
|
8.614
|
NM_001195626
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr2_-_206950895
|
8.603
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr12_+_122459791
|
8.601
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr22_+_45705766
|
8.505
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr2_+_219575507
|
8.497
|
NM_014640
|
TTLL4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr5_-_148033715
|
8.396
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr1_+_60280462
|
8.395
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr7_-_27205148
|
8.385
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr19_+_50529211
|
8.376
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr16_+_18995255
|
8.367
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr9_-_77503009
|
8.329
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr5_+_56110899
|
8.128
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr1_-_146644045
|
8.123
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr1_-_23857711
|
8.076
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr17_+_8214139
|
8.044
|
NM_025014
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr1_+_113615787
|
7.871
|
NM_014813
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr8_-_48650703
|
7.821
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr11_+_45868956
|
7.804
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr7_+_20370724
|
7.753
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr20_-_48532067
|
7.638
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr2_+_128848752
|
7.629
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr1_-_204121144
|
7.615
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chr1_-_202679415
|
7.602
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr1_-_184724021
|
7.598
|
NM_025191
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr16_+_18995508
|
7.556
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr12_+_49372235
|
7.554
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr4_-_170947428
|
7.547
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr13_+_49549953
|
7.485
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr4_+_75858205
|
7.483
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr1_-_169863048
|
7.464
|
NM_020423
NM_181093
|
SCYL3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr6_-_26659913
|
7.432
|
NM_001242797
NM_001242798
NM_001242799
NM_024639
|
ZNF322
|
zinc finger protein 322
|
|
chr20_-_30795346
|
7.420
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr8_+_28351694
|
7.417
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr6_+_154407641
|
7.415
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr17_-_7197866
|
7.395
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr12_-_42538432
|
7.334
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr6_-_35464716
|
7.330
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr12_+_54366909
|
7.314
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr9_-_72287097
|
7.289
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr8_-_122653493
|
7.266
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr1_+_203274646
|
7.159
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr12_-_102224509
|
7.029
|
NM_024312
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits
|
|
chr5_-_74326723
|
7.028
|
NM_016591
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chrX_+_152907892
|
7.011
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr12_-_57400199
|
6.999
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr9_-_27529434
|
6.977
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr11_-_86666211
|
6.928
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr16_-_2908105
|
6.910
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr19_-_9929682
|
6.898
|
NM_017703
|
FBXL12
|
F-box and leucine-rich repeat protein 12
|
|
chr22_+_25960738
|
6.877
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr15_-_45815001
|
6.875
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr16_+_640056
|
6.770
|
NM_001172665
NM_001172666
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr12_-_42631982
|
6.743
|
NM_001190977
|
YAF2
|
YY1 associated factor 2
|
|
chr4_-_1166959
|
6.734
|
NM_001128325
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr20_+_55966440
|
6.723
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr13_-_52585472
|
6.701
|
NM_000053
NM_001005918
NM_001243182
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr12_+_6421816
|
6.692
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chrX_+_16804531
|
6.650
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr1_+_204485461
|
6.645
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr13_+_48877775
|
6.615
|
NM_000321
|
RB1
|
retinoblastoma 1
|
|
chr4_+_56212381
|
6.590
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr22_-_39636906
|
6.588
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr5_+_157170702
|
6.568
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr12_+_103981050
|
6.524
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr3_-_169899519
|
6.503
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr4_+_88571453
|
6.475
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr19_+_46806855
|
6.441
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr5_+_49962771
|
6.439
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr2_-_242255114
|
6.433
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr9_-_35115844
|
6.413
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr17_+_29718641
|
6.392
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_+_56815036
|
6.328
|
NM_025009
|
CEP135
|
centrosomal protein 135kDa
|
|
chr17_+_41476351
|
6.318
|
NM_001661
|
ARL4D
|
ADP-ribosylation factor-like 4D
|
|
chr9_+_99212379
|
6.317
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr9_-_77502635
|
6.292
|
NM_001177310
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr9_-_129884998
|
6.270
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr13_+_39612440
|
6.265
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr3_-_101039403
|
6.245
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr4_-_1202749
|
6.232
|
NM_001199021
|
LOC100130872
SPON2
|
uncharacterized LOC100130872
spondin 2, extracellular matrix protein
|
|
chr1_-_115632047
|
6.209
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr8_-_37823986
|
6.208
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr12_+_111843743
|
6.193
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr5_+_131993864
|
6.170
|
NM_002188
|
IL13
|
interleukin 13
|
|
chr12_+_6420098
|
6.156
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr11_-_62389571
|
6.148
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr8_-_67579446
|
6.136
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr15_+_45879416
|
6.127
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr4_-_1166547
|
6.108
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr5_+_42550181
|
6.072
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr10_+_101419169
|
6.045
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr22_+_45704821
|
6.008
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr2_-_214016332
|
6.001
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr19_-_36980336
|
5.957
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr19_+_40973049
|
5.943
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr1_-_118472205
|
5.933
|
NM_001135589
NM_017686
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr12_-_69357019
|
5.920
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr19_+_34287662
|
5.901
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr9_-_36400817
|
5.886
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr5_+_161112657
|
5.875
|
NM_000811
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6
|
|
chr17_+_32597239
|
5.872
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr12_-_114846191
|
5.849
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr15_-_29862926
|
5.798
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr1_+_29213569
|
5.792
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr11_+_45868668
|
5.779
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr5_-_159739482
|
5.769
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr10_-_50747080
|
5.746
|
NM_000124
|
ERCC6
PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6
piggyBac transposable element derived 3
|
|
chrX_+_21857655
|
5.734
|
NM_015884
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr6_+_138188352
|
5.730
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_+_172628157
|
5.716
|
NM_000639
|
FASLG
|
Fas ligand (TNF superfamily, member 6)
|
|
chr8_-_144623611
|
5.708
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr17_+_77751929
|
5.698
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr8_+_121137297
|
5.684
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr14_-_88459469
|
5.657
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chr13_-_52733669
|
5.641
|
NM_152720
|
NEK3
|
NIMA (never in mitosis gene a)-related kinase 3
|
|
chr17_+_61699786
|
5.637
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_+_14075861
|
5.636
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr5_-_32313069
|
5.619
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr1_-_182573393
|
5.610
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr3_+_183353355
|
5.572
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chrX_-_15353675
|
5.567
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr8_+_22102618
|
5.545
|
NM_001722
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|
|
chr17_+_8213555
|
5.540
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr9_+_101867401
|
5.523
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr6_+_106534188
|
5.469
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_106546736
|
5.455
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr16_+_639356
|
5.451
|
NM_001172663
|
RAB40C
|
RAB40C, member RAS oncogene family
|