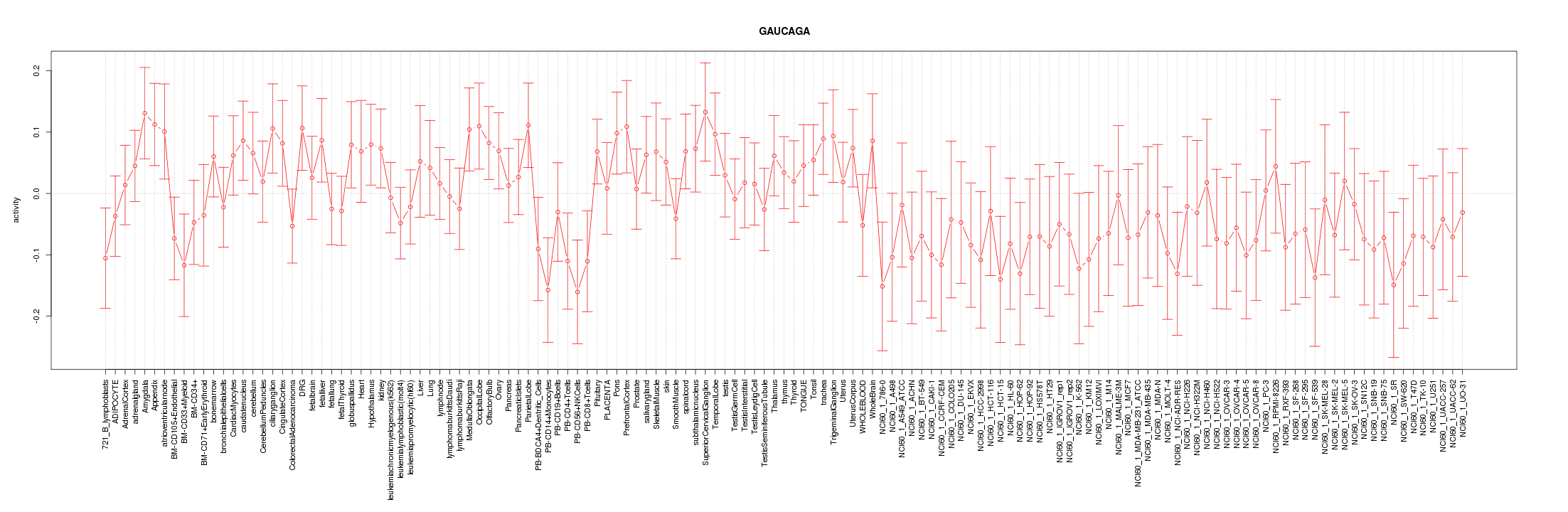
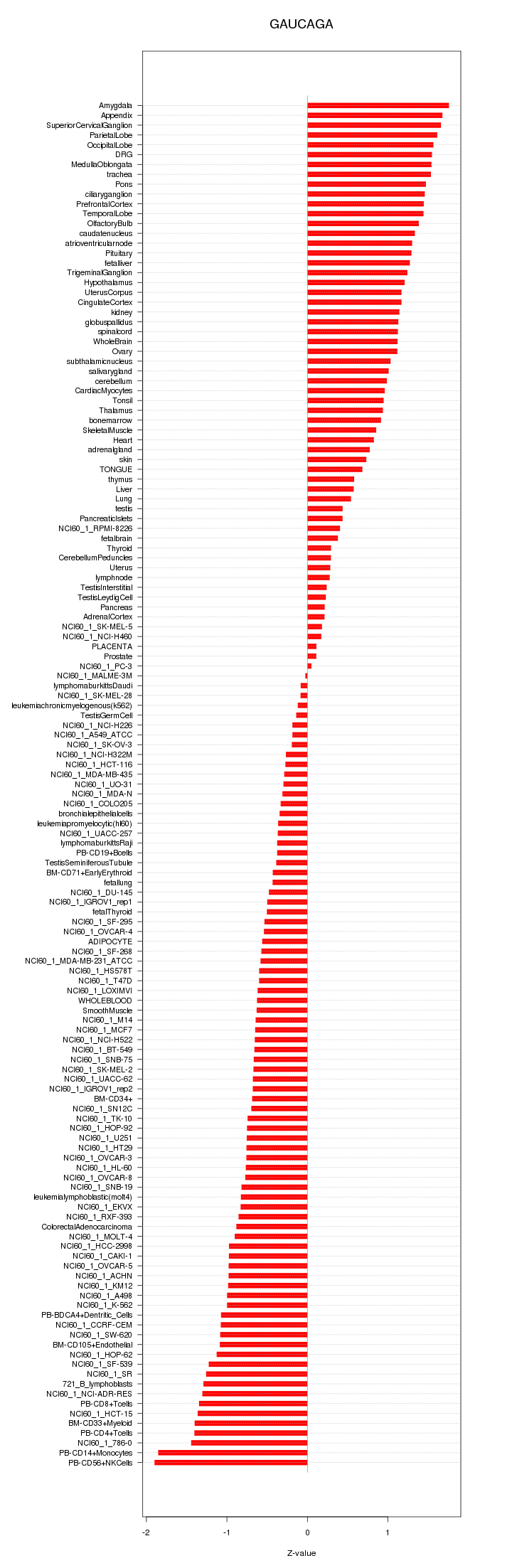
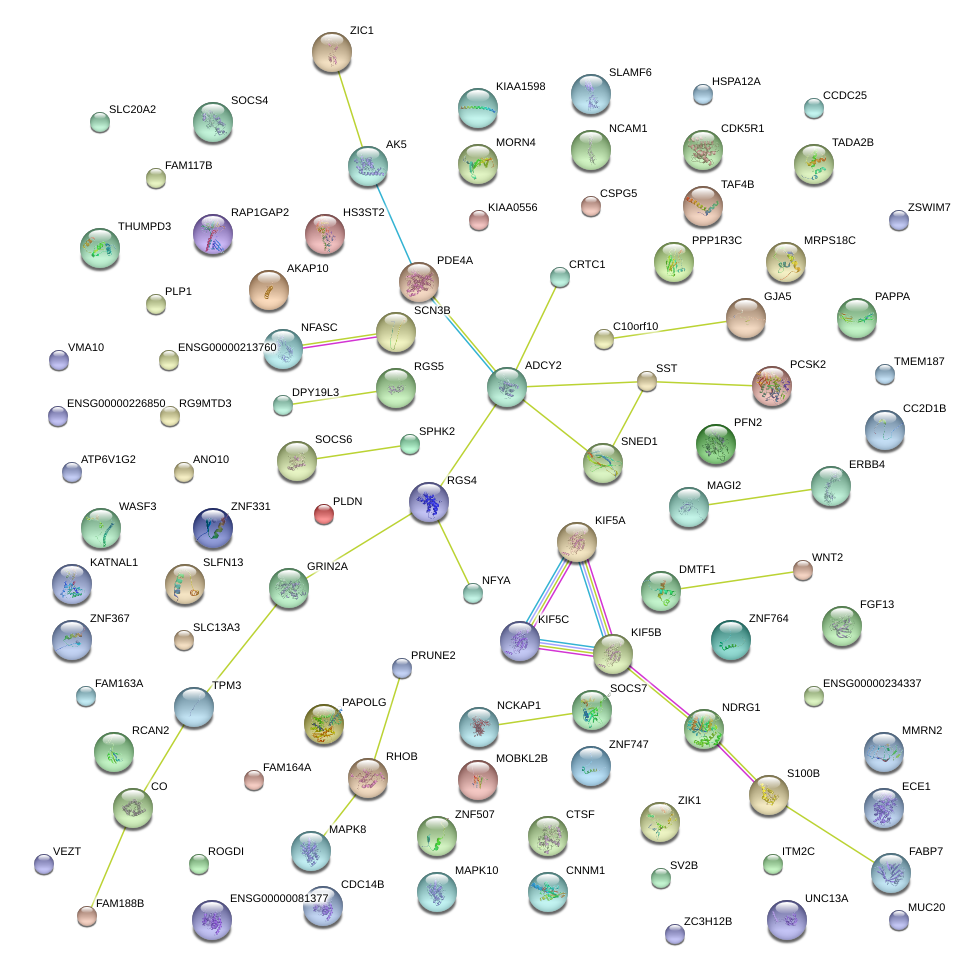
|
chr15_+_91643429
|
9.603
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr15_+_91643181
|
8.352
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr10_-_118501981
|
6.013
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chrX_+_64708614
|
5.910
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr11_-_66335958
|
5.809
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chrX_+_103031433
|
5.260
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr13_-_30881141
|
5.226
|
NM_001014380
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chrX_+_103031753
|
4.963
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr6_-_46293628
|
4.922
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr13_-_30881556
|
4.901
|
NM_032116
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr17_-_15902907
|
4.836
|
NM_001042697
NM_001042698
|
ZSWIM7
|
zinc finger, SWIM-type containing 7
|
|
chr11_-_123525300
|
4.770
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr19_+_58095507
|
4.765
|
NM_001010879
|
ZIK1
|
zinc finger protein interacting with K protein 1 homolog (mouse)
|
|
chr2_+_203499819
|
4.630
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr21_-_48024985
|
4.464
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr19_+_54024176
|
4.322
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr2_-_183903225
|
4.291
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr2_+_149632772
|
4.118
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr9_-_79520878
|
3.765
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr19_+_10541334
|
3.675
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr9_-_27529434
|
3.672
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr19_+_54041332
|
3.560
|
NM_001079906
NM_001253798
NM_001253799
|
ZNF331
|
zinc finger protein 331
|
|
chr10_-_93392857
|
3.488
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr19_+_18794391
|
3.401
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr19_+_49122547
|
3.354
|
NM_001204158
NM_001243876
NM_020126
|
SPHK2
|
sphingosine kinase 2
|
|
chr6_+_41040687
|
3.250
|
NM_002505
NM_021705
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr17_+_2699731
|
3.249
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr1_-_163172624
|
3.235
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr2_+_20646831
|
3.234
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr19_+_49122789
|
3.223
|
NM_001204159
|
SPHK2
|
sphingosine kinase 2
|
|
chr1_-_163172863
|
3.119
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr6_-_31514620
|
3.108
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr19_+_32896515
|
3.097
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr20_-_45313122
|
3.030
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr7_+_30960914
|
3.025
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr1_-_52831817
|
3.025
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr1_+_179712297
|
3.023
|
NM_173509
|
FAM163A
|
family with sequence similarity 163, member A
|
|
chr3_-_187388105
|
3.021
|
NM_001048
|
SST
|
somatostatin
|
|
chr16_-_30546145
|
2.960
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chrX_-_137793826
|
2.938
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chrX_-_137949716
|
2.920
|
NM_001139502
|
FGF13
|
fibroblast growth factor 13
|
|
chr17_-_19880991
|
2.913
|
NM_007202
|
AKAP10
|
A kinase (PRKA) anchor protein 10
|
|
chr20_-_45280059
|
2.910
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr16_+_22825806
|
2.845
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr19_+_54058493
|
2.827
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr17_-_33775761
|
2.824
|
NM_144682
|
SLFN13
|
schlafen family member 13
|
|
chr1_+_77748286
|
2.792
|
NM_012093
|
AK5
|
adenylate kinase 5
|
|
chr3_+_195447751
|
2.771
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr3_+_147127151
|
2.733
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr1_-_154155698
|
2.710
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr13_+_27131799
|
2.708
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr9_-_99382073
|
2.705
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr16_-_10276610
|
2.687
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr1_+_163038395
|
2.675
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr16_-_4852571
|
2.632
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr8_-_42397048
|
2.609
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr8_-_27630096
|
2.597
|
NM_018246
|
CCDC25
|
coiled-coil domain containing 25
|
|
chr10_-_45474210
|
2.593
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chrX_-_138287180
|
2.590
|
NM_001139498
NM_001139500
NM_001139501
|
FGF13
|
fibroblast growth factor 13
|
|
chr10_+_101088794
|
2.555
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr1_+_204913353
|
2.546
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr10_-_99393175
|
2.515
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr10_-_88717319
|
2.503
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr4_+_84377117
|
2.495
|
NM_016067
|
MRPS18C
|
mitochondrial ribosomal protein S18C
|
|
chr3_-_47620186
|
2.455
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr2_-_213403280
|
2.454
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr7_+_30951414
|
2.428
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr7_-_116963328
|
2.410
|
NM_003391
|
WNT2
|
wingless-type MMTV integration site family member 2
|
|
chr9_+_37753769
|
2.396
|
NM_144964
|
RG9MTD3
|
RNA (guanine-9-) methyltransferase domain containing 3
|
|
chr1_-_21616888
|
2.377
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_+_163041694
|
2.358
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr8_+_79578258
|
2.354
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr2_-_200820458
|
2.316
|
NM_001039693
|
TYW5
|
tRNA-yW synthesizing protein 5
|
|
chr5_+_7396030
|
2.295
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr10_-_118765087
|
2.280
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr8_-_134309448
|
2.271
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr1_-_147232684
|
2.264
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr17_+_36507708
|
2.257
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chrX_+_153237990
|
2.249
|
NM_003492
|
TMEM187
|
transmembrane protein 187
|
|
chr1_+_77747602
|
2.239
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr11_+_112831968
|
2.231
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr3_+_9404706
|
2.227
|
NM_001114092
NM_015453
|
THUMPD3
|
THUMP domain containing 3
|
|
chr2_+_241938254
|
2.222
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr16_+_27561441
|
2.209
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr4_-_87028805
|
2.191
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr18_+_23806385
|
2.188
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr10_-_99393851
|
2.157
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr19_+_32836500
|
2.139
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr2_+_60983354
|
2.137
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr20_+_17207596
|
2.137
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr7_-_79082870
|
2.137
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr2_+_231729501
|
2.127
|
NM_001012514
NM_001012516
NM_030926
|
ITM2C
|
integral membrane protein 2C
|
|
chr15_+_45879416
|
2.121
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr1_+_163038934
|
2.116
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr19_-_17799005
|
2.105
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr7_+_86781869
|
2.087
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr1_-_160492982
|
2.081
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr17_+_30813928
|
2.079
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr3_-_149688638
|
2.058
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr6_+_123100645
|
2.057
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr4_+_7045142
|
2.052
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr12_+_95611514
|
2.048
|
NM_017599
|
VEZT
|
vezatin, adherens junctions transmembrane protein
|
|
chr2_-_103353300
|
2.048
|
NM_032718
|
MFSD9
|
major facilitator superfamily domain containing 9
|
|
chr2_-_10830102
|
2.036
|
NM_024894
|
NOL10
|
nucleolar protein 10
|
|
chr12_+_4918341
|
2.027
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr9_-_23821842
|
2.024
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_-_135194823
|
2.022
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr8_+_120885899
|
2.013
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr13_-_46626845
|
2.013
|
NM_015070
|
ZC3H13
|
zinc finger CCCH-type containing 13
|
|
chr19_-_37407079
|
2.010
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr5_-_58334936
|
2.008
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_-_136871956
|
2.004
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr16_-_10276115
|
1.994
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr20_+_17206751
|
1.983
|
NM_001201528
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr22_-_36013303
|
1.971
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr4_-_25032306
|
1.948
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr11_+_12399024
|
1.941
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr1_+_24882565
|
1.939
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr3_+_101443436
|
1.927
|
NM_024548
|
CEP97
|
centrosomal protein 97kDa
|
|
chr3_+_32148000
|
1.919
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr6_+_154407641
|
1.918
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr1_+_163038564
|
1.913
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr8_-_21966812
|
1.903
|
NM_024815
|
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18
|
|
chr3_-_171528227
|
1.902
|
NM_001130081
NM_002662
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific
|
|
chr17_-_74236272
|
1.896
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr4_-_87374282
|
1.883
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr8_+_42752019
|
1.883
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr7_+_108210188
|
1.879
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr1_-_114354906
|
1.874
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr19_+_2328628
|
1.869
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr4_+_62362838
|
1.869
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr3_+_132379130
|
1.868
|
NM_024818
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr5_-_118324223
|
1.863
|
NM_173666
|
DTWD2
|
DTW domain containing 2
|
|
chr11_+_45868956
|
1.859
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr2_-_152830448
|
1.855
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr22_-_36019363
|
1.851
|
NM_203377
|
MB
|
myoglobin
|
|
chr7_+_86781644
|
1.849
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr3_-_9291251
|
1.840
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr20_+_32077904
|
1.840
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr16_-_1020981
|
1.836
|
NM_022773
|
LMF1
|
lipase maturation factor 1
|
|
chr7_-_44924946
|
1.820
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr4_+_95679075
|
1.820
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr12_+_11081833
|
1.815
|
NM_005042
NM_001110213
|
PRH2
|
proline-rich protein HaeIII subfamily 2
|
|
chr13_-_22033505
|
1.813
|
NM_153251
|
ZDHHC20
|
zinc finger, DHHC-type containing 20
|
|
chr22_-_50523780
|
1.810
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr18_-_28622624
|
1.789
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr8_-_17658385
|
1.784
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr18_-_78005230
|
1.780
|
NM_032510
|
PARD6G
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr1_+_151032150
|
1.779
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_-_97321094
|
1.778
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chrX_+_57618268
|
1.767
|
NM_007157
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr9_+_103947316
|
1.765
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr1_-_40157067
|
1.759
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr12_+_133562941
|
1.754
|
NM_019591
|
ZNF26
|
zinc finger protein 26
|
|
chr1_-_21616648
|
1.752
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr22_+_50247480
|
1.751
|
NM_014838
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr1_-_202679415
|
1.750
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr19_+_57019196
|
1.750
|
NM_020813
|
ZNF471
|
zinc finger protein 471
|
|
chr19_+_48949029
|
1.744
|
NM_031485
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr18_-_56940616
|
1.738
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr6_-_132272311
|
1.738
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr16_+_89284084
|
1.735
|
NM_001201407
NM_182531
|
ZNF778
|
zinc finger protein 778
|
|
chr12_-_133532847
|
1.725
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr11_-_118661898
|
1.721
|
NM_004397
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr11_-_108093328
|
1.714
|
NM_002519
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr9_+_101705710
|
1.713
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr2_-_2334887
|
1.705
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_+_27215295
|
1.697
|
NM_001145348
|
JMJD5
|
jumonji domain containing 5
|
|
chr11_+_111473099
|
1.688
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr12_-_124457099
|
1.676
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr1_-_202612580
|
1.674
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr2_+_219824346
|
1.672
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr19_+_46800296
|
1.667
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr11_+_45868668
|
1.659
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr3_-_127541975
|
1.658
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr12_+_78225068
|
1.653
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_-_179169330
|
1.649
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr19_-_58400287
|
1.647
|
NM_001144989
|
ZNF814
|
zinc finger protein 814
|
|
chrX_+_15756391
|
1.636
|
NM_007220
|
CA5B
|
carbonic anhydrase VB, mitochondrial
|
|
chr1_-_28415071
|
1.628
|
NM_001990
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr12_-_42538432
|
1.627
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr21_+_27107257
|
1.619
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr16_+_27214806
|
1.605
|
NM_024773
|
JMJD5
|
jumonji domain containing 5
|
|
chr9_+_17134974
|
1.604
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr6_-_168476531
|
1.595
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr2_-_152955208
|
1.587
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr1_-_151798552
|
1.583
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr19_+_52772823
|
1.582
|
NM_001010851
|
ZNF766
|
zinc finger protein 766
|
|
chr4_-_87281374
|
1.582
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr10_-_21463019
|
1.581
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr15_-_93198602
|
1.579
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr10_+_104629209
|
1.575
|
NM_020682
|
AS3MT
|
arsenic (+3 oxidation state) methyltransferase
|
|
chr3_-_57199402
|
1.575
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr22_-_34316325
|
1.568
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr16_+_57406398
|
1.565
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr3_+_108021331
|
1.561
|
NM_007072
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr19_-_48867149
|
1.558
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr20_-_4229519
|
1.547
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr16_-_89556886
|
1.545
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr10_+_14920781
|
1.540
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|