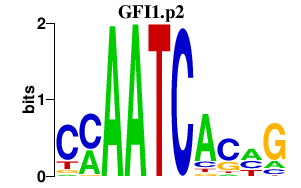
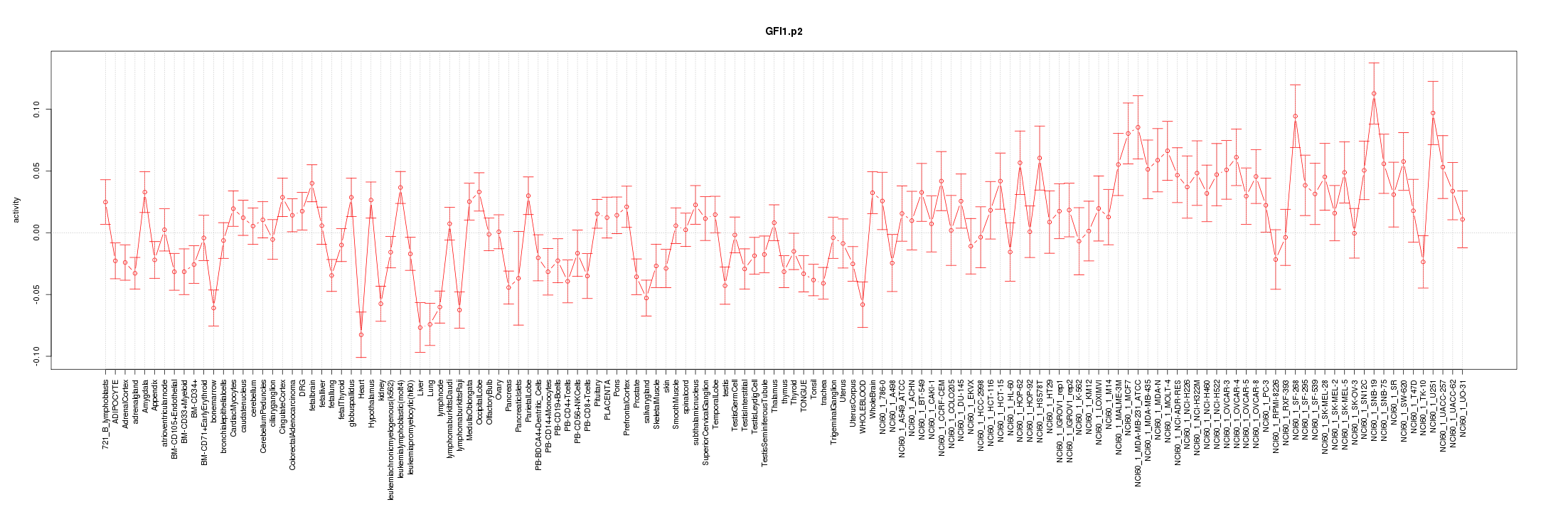
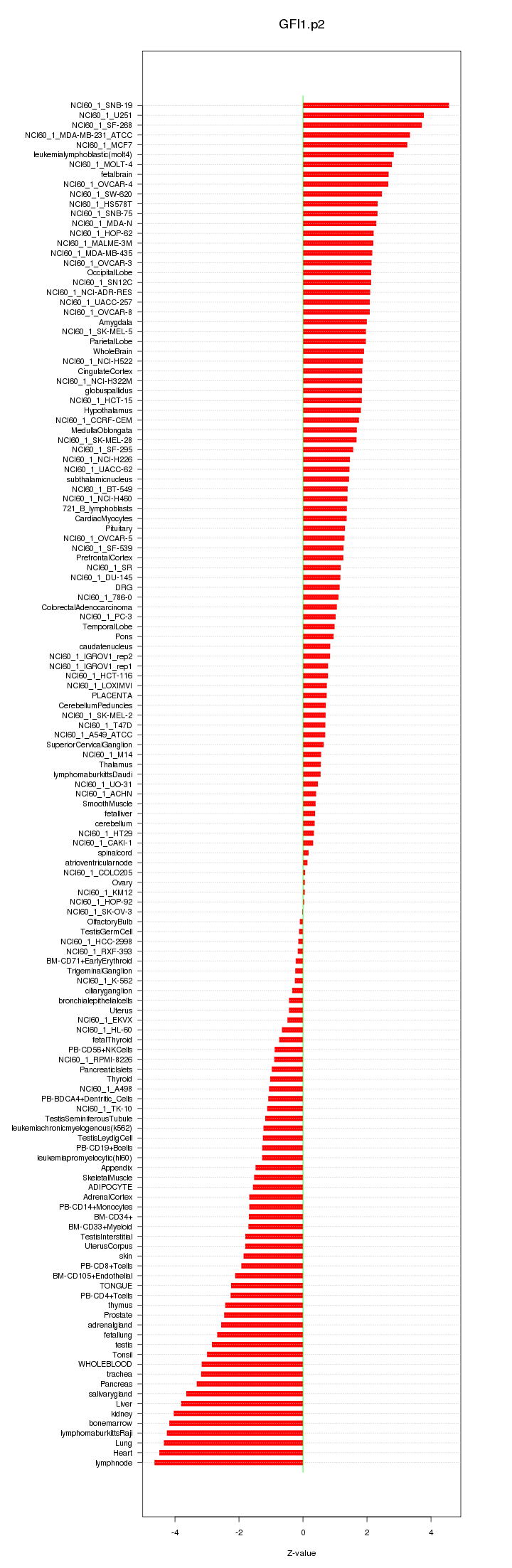
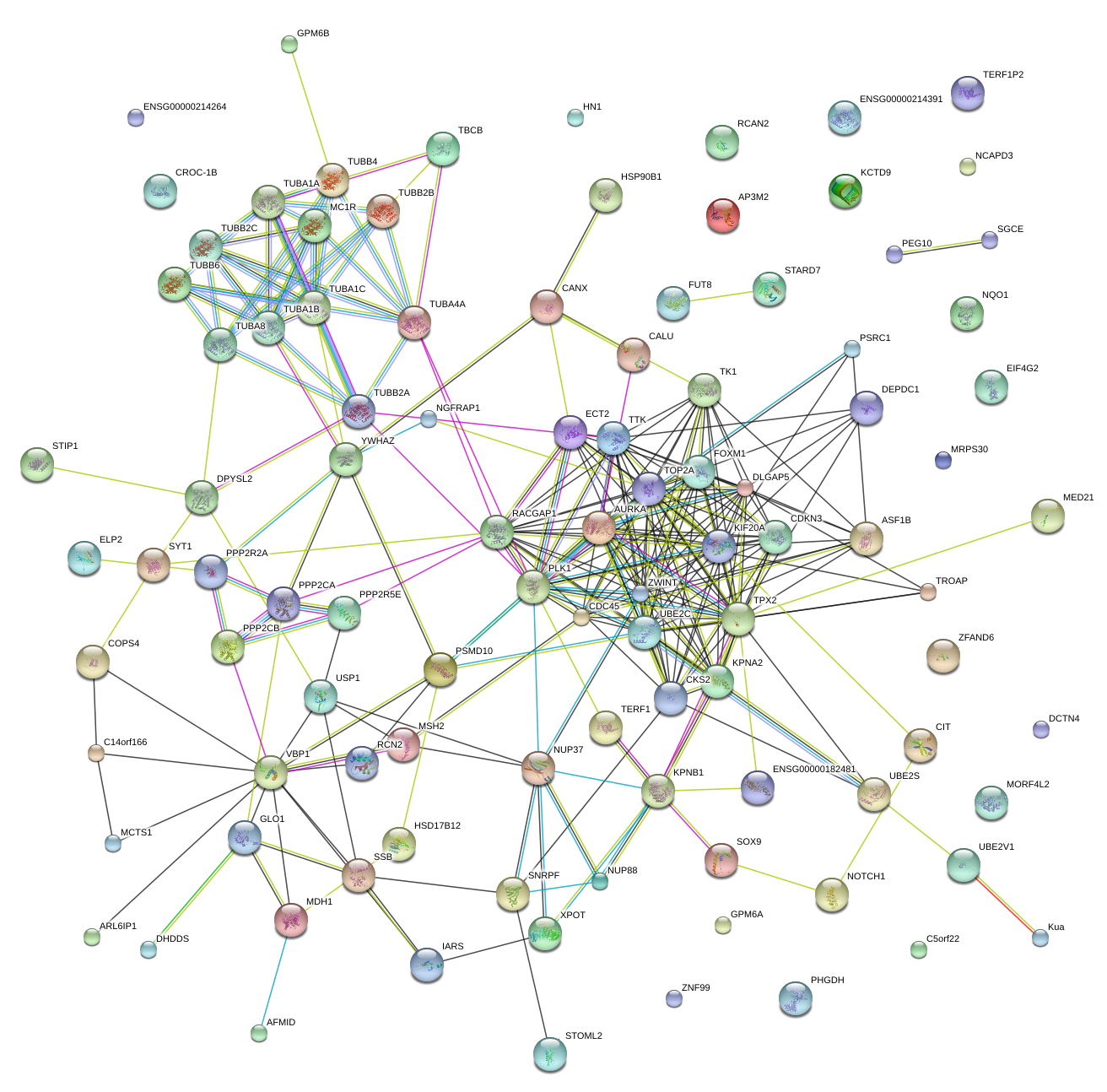
|
chr3_+_172468469
|
33.767
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr19_-_14247267
|
28.288
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr19_-_14247400
|
26.128
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr9_+_91926109
|
23.486
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr14_-_55658205
|
23.014
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr17_-_38574042
|
22.942
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr12_-_120315074
|
19.586
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr12_+_104324157
|
19.554
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr8_-_25315941
|
18.777
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chrX_-_107334749
|
18.737
|
NM_002814
NM_170750
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10
|
|
chrX_+_154444700
|
18.343
|
NM_003372
|
VBP1
|
von Hippel-Lindau binding protein 1
|
|
chr12_+_104324192
|
17.652
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr1_+_62902325
|
17.468
|
NM_003368
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr17_-_76183074
|
17.451
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr8_-_101964248
|
16.639
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr8_-_101964247
|
16.516
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr12_+_104324214
|
16.437
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr8_-_101964268
|
16.354
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr8_-_101964236
|
16.216
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr12_+_104324186
|
16.037
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr8_-_101964258
|
15.775
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr16_-_69760354
|
15.480
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr8_-_101964197
|
15.444
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr1_-_68962679
|
15.249
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr6_-_38670882
|
15.115
|
|
GLO1
|
glyoxalase I
|
|
chr6_-_38670904
|
14.746
|
|
GLO1
|
glyoxalase I
|
|
chr2_+_47630291
|
14.678
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr12_-_102513846
|
14.552
|
|
NUP37
|
nucleoporin 37kDa
|
|
chr6_-_38670922
|
14.423
|
NM_006708
|
GLO1
|
glyoxalase I
|
|
chr2_+_47630190
|
14.397
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chrX_+_119737905
|
14.352
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_47630246
|
14.292
|
NM_000251
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chrX_+_119737984
|
14.275
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr14_+_54863672
|
14.274
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr12_+_96252770
|
14.040
|
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr12_+_64803741
|
14.014
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr12_+_96252705
|
13.834
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr16_+_23690160
|
13.655
|
|
PLK1
|
polo-like kinase 1
|
|
chr16_+_23690212
|
13.558
|
|
PLK1
|
polo-like kinase 1
|
|
chr6_-_38670829
|
13.045
|
|
GLO1
|
glyoxalase I
|
|
chr16_+_23690234
|
12.773
|
|
PLK1
|
polo-like kinase 1
|
|
chrX_-_13835012
|
12.646
|
|
GPM6B
|
glycoprotein M6B
|
|
chr16_+_23690191
|
12.589
|
NM_005030
|
PLK1
|
polo-like kinase 1
|
|
chr16_+_89989686
|
12.527
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr20_-_54967221
|
12.413
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr16_+_23690190
|
12.110
|
|
PLK1
|
polo-like kinase 1
|
|
chr14_-_64010087
|
11.925
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr19_-_55919324
|
11.846
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr8_+_73921061
|
11.709
|
NM_003218
NM_017489
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr17_+_70117168
|
11.545
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr2_+_170662044
|
11.495
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr8_-_101964334
|
11.370
|
NM_145690
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr17_+_76183397
|
11.345
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr19_-_55918980
|
11.338
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr14_-_64009971
|
11.307
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr17_+_70117177
|
11.291
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr5_+_137514416
|
11.151
|
NM_005733
|
KIF20A
|
kinesin family member 20A
|
|
chr1_-_68962726
|
11.133
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr16_-_69760367
|
11.091
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr16_-_69760444
|
11.030
|
NM_000903
NM_001025433
NM_001025434
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr12_+_79258588
|
10.943
|
|
SYT1
|
synaptotagmin I
|
|
chr11_-_10830452
|
10.770
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr4_-_176734181
|
10.738
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr12_+_49716970
|
10.726
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr5_+_137514753
|
10.674
|
|
KIF20A
|
kinesin family member 20A
|
|
chr8_-_25315791
|
10.535
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr17_+_45727274
|
10.533
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr15_+_80364902
|
10.360
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr1_-_109825767
|
10.248
|
NM_001005290
NM_001032291
NM_032636
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr7_-_94285477
|
10.196
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr19_+_36606337
|
10.196
|
|
TBCB
|
tubulin folding cofactor B
|
|
chr8_+_26150731
|
10.063
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr20_+_30326903
|
10.058
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr8_+_42010463
|
9.947
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr14_+_65878579
|
9.919
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr1_-_109825683
|
9.917
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr12_-_2986299
|
9.877
|
NM_001243088
NM_001243089
NM_021953
NM_202002
NM_202003
|
FOXM1
|
forkhead box M1
|
|
chr12_-_50419160
|
9.852
|
NM_001126103
NM_001126104
NM_013277
|
RACGAP1
|
Rac GTPase activating protein 1
|
|
chr2_+_63815742
|
9.840
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr7_+_94285676
|
9.835
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr2_+_63816092
|
9.832
|
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr14_-_64010063
|
9.822
|
NM_006246
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr7_-_94285426
|
9.790
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr15_+_77223942
|
9.763
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr2_+_63816143
|
9.730
|
NM_001199111
NM_001199112
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr11_+_63953630
|
9.565
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr7_-_94285372
|
9.534
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr8_+_26435339
|
9.476
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_+_102631267
|
9.445
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr17_-_5322870
|
9.438
|
|
NUP88
|
nucleoporin 88kDa
|
|
chr20_+_44441334
|
9.430
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr17_-_73150769
|
9.312
|
NM_001002032
NM_001002033
NM_016185
|
HN1
|
hematological and neurological expressed 1
|
|
chr17_+_70117154
|
9.289
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr5_-_150138081
|
9.280
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr11_+_43702142
|
9.232
|
NM_016142
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12
|
|
chr7_+_94285736
|
9.229
|
|
PEG10
|
paternally expressed 10
|
|
chr12_+_27175482
|
9.220
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr12_-_49582800
|
9.211
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr11_-_134093869
|
9.073
|
|
NCAPD3
|
non-SMC condensin II complex, subunit D3
|
|
chr2_-_96874507
|
8.974
|
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr9_-_35103089
|
8.972
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr5_-_133561745
|
8.955
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr5_+_44808967
|
8.943
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr17_+_66031814
|
8.903
|
NM_002266
|
KPNA2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1)
|
|
chr19_-_22966972
|
8.878
|
NM_001080409
|
ZNF99
|
zinc finger protein 99
|
|
chr5_+_31532372
|
8.847
|
NM_018356
|
C5orf22
|
chromosome 5 open reading frame 22
|
|
chrX_-_102941582
|
8.845
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr2_-_96873938
|
8.827
|
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr10_-_58121011
|
8.802
|
|
ZWINT
|
ZW10 interactor
|
|
chr16_-_18812691
|
8.753
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr10_-_58121019
|
8.735
|
NM_001005413
NM_007057
NM_032997
|
ZWINT
|
ZW10 interactor
|
|
chr14_+_52456322
|
8.714
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr20_+_44441247
|
8.627
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chrX_+_102631408
|
8.614
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr11_-_10830299
|
8.581
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr5_-_133561551
|
8.569
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr6_+_80714321
|
8.539
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr22_+_19467467
|
8.419
|
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr6_-_38670871
|
8.419
|
|
GLO1
|
glyoxalase I
|
|
chr6_-_38670915
|
8.408
|
|
GLO1
|
glyoxalase I
|
|
chr5_+_179132676
|
8.401
|
|
CANX
|
calnexin
|
|
chr17_+_66031885
|
8.339
|
|
KPNA2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1)
|
|
chr7_+_128379452
|
8.333
|
|
CALU
|
calumenin
|
|
chr20_-_48770302
|
8.333
|
NM_001162505
NM_199129
NM_199203
|
TMEM189
TMEM189-UBE2V1
|
transmembrane protein 189
TMEM189-UBE2V1 readthrough
|
|
chr9_-_35103048
|
8.312
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr11_-_134093826
|
8.292
|
|
NCAPD3
|
non-SMC condensin II complex, subunit D3
|
|
chr9_-_95055966
|
8.274
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr1_+_120254418
|
8.243
|
NM_006623
|
PHGDH
|
phosphoglycerate dehydrogenase
|
|
chr9_-_95055986
|
8.242
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr1_+_62902653
|
8.203
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr7_+_128379400
|
8.191
|
|
CALU
|
calumenin
|
|
chr4_+_83956238
|
8.165
|
NM_016129
|
COPS4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis)
|
|
chr1_+_62902735
|
8.159
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr9_-_35103144
|
8.145
|
NM_013442
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr7_+_128379428
|
8.136
|
|
CALU
|
calumenin
|
|
chr11_+_63953586
|
8.097
|
NM_006819
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr11_-_119293848
|
8.085
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr1_+_62902708
|
8.022
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr22_+_19467413
|
8.018
|
NM_001178010
NM_001178011
NM_003504
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr17_+_6347733
|
8.016
|
NM_001195228
NM_019013
|
FAM64A
|
family with sequence similarity 64, member A
|
|
chr12_-_2986095
|
8.004
|
|
FOXM1
|
forkhead box M1
|
|
chr9_+_131446105
|
7.998
|
|
SET
|
SET nuclear oncogene
|
|
chr8_-_124408665
|
7.961
|
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr7_+_94285620
|
7.941
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr11_-_10830581
|
7.939
|
NM_001042559
NM_001418
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr17_+_34842472
|
7.907
|
NM_004773
|
ZNHIT3
|
zinc finger, HIT-type containing 3
|
|
chr1_-_31769599
|
7.846
|
NM_004814
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5)
|
|
chr15_-_83876769
|
7.833
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr20_+_44441671
|
7.806
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr6_+_123100645
|
7.798
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr15_-_42749710
|
7.788
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr1_-_95392449
|
7.772
|
|
CNN3
|
calponin 3, acidic
|
|
chr11_-_71159442
|
7.749
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr9_+_36190885
|
7.706
|
|
CLTA
|
clathrin, light chain A
|
|
chr5_-_137878808
|
7.675
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr1_-_150669499
|
7.648
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr2_-_234763182
|
7.648
|
NM_018410
|
HJURP
|
Holliday junction recognition protein
|
|
chr12_+_6603258
|
7.636
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr2_-_234763145
|
7.634
|
|
HJURP
|
Holliday junction recognition protein
|
|
chr7_+_128379395
|
7.624
|
|
CALU
|
calumenin
|
|
chrX_+_119738542
|
7.531
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr20_+_47662782
|
7.527
|
NM_001316
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr3_-_145879222
|
7.510
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr17_-_40075153
|
7.493
|
|
ACLY
|
ATP citrate lyase
|
|
chr10_+_62538250
|
7.481
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr5_-_137090037
|
7.475
|
NM_006805
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr6_-_18264638
|
7.439
|
NM_001134709
NM_003472
|
DEK
|
DEK oncogene
|
|
chr1_+_214776494
|
7.437
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr12_+_6603291
|
7.421
|
NM_014865
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr3_+_50273745
|
7.421
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr19_+_36605887
|
7.420
|
NM_001281
|
TBCB
|
tubulin folding cofactor B
|
|
chr6_+_111135823
|
7.420
|
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr9_+_131445933
|
7.383
|
NM_001122821
|
SET
|
SET nuclear oncogene
|
|
chr11_+_63953666
|
7.355
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr17_-_5322982
|
7.351
|
NM_002532
|
NUP88
|
nucleoporin 88kDa
|
|
chr8_-_124408688
|
7.344
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr12_+_56618104
|
7.312
|
NM_024068
|
OBFC2B
|
oligonucleotide/oligosaccharide-binding fold containing 2B
|
|
chr2_-_96874569
|
7.306
|
NM_020151
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr4_-_174255454
|
7.272
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr16_-_81129882
|
7.269
|
NM_004483
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier)
|
|
chr8_-_124408458
|
7.265
|
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chrX_+_119737743
|
7.261
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr12_+_56618150
|
7.176
|
|
OBFC2B
|
oligonucleotide/oligosaccharide-binding fold containing 2B
|
|
chr5_-_31532164
|
7.173
|
NM_001100412
NM_013235
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr6_+_33359620
|
7.168
|
|
KIFC1
|
kinesin family member C1
|
|
chr8_+_66556887
|
7.162
|
NM_001145839
NM_014637
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chr12_+_79258517
|
7.155
|
|
SYT1
|
synaptotagmin I
|
|
chrX_-_37706816
|
7.154
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr1_-_33282844
|
7.118
|
|
YARS
|
tyrosyl-tRNA synthetase
|
|
chr6_+_33359604
|
7.084
|
|
KIFC1
|
kinesin family member C1
|
|
chrX_+_48380163
|
7.071
|
NM_006579
|
EBP
|
emopamil binding protein (sterol isomerase)
|
|
chr2_-_47403652
|
7.053
|
|
CALM2
|
calmodulin 2 (phosphorylase kinase, delta)
|
|
chr1_-_33283632
|
7.031
|
NM_003680
|
YARS
|
tyrosyl-tRNA synthetase
|
|
chr17_+_45727558
|
7.020
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chrX_-_102319080
|
6.986
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr5_+_179125977
|
6.977
|
|
CANX
|
calnexin
|
|
chr4_-_76598611
|
6.963
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr14_-_105487413
|
6.942
|
|
CDCA4
|
cell division cycle associated 4
|
|
chr2_+_9346909
|
6.904
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr4_+_166248816
|
6.890
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|