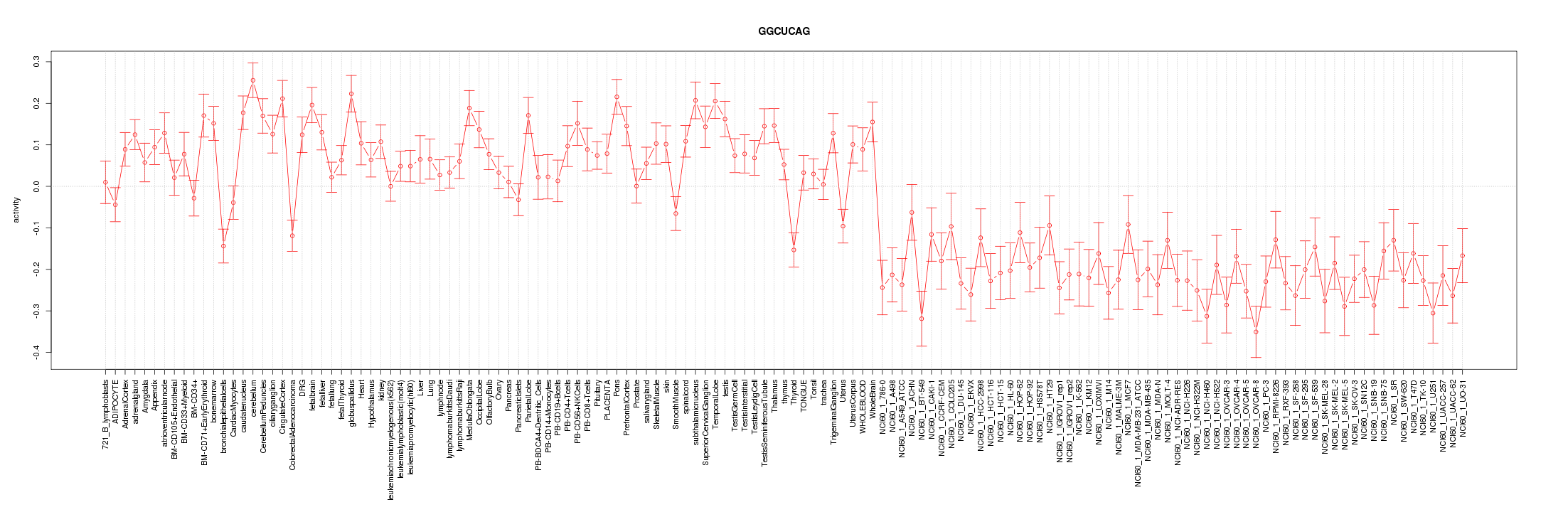
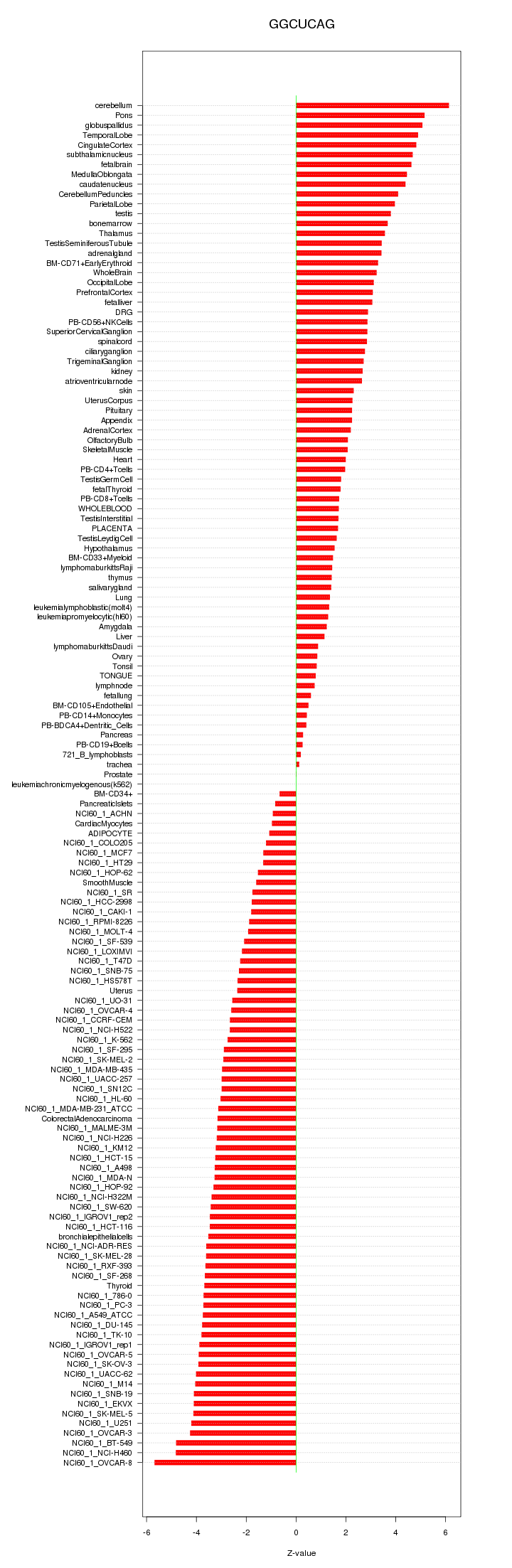
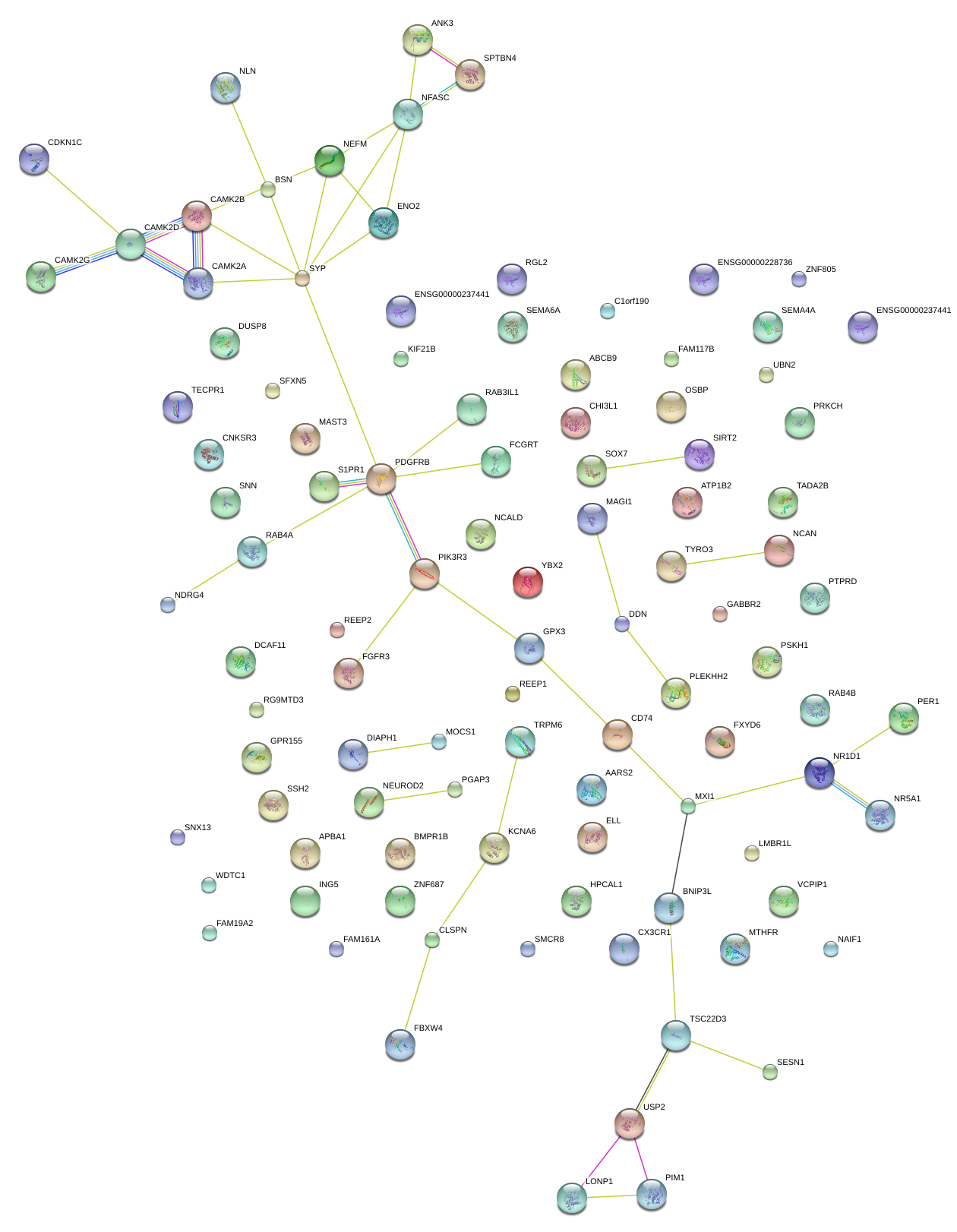
|
chr8_+_24771268
|
66.159
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_24772454
|
61.293
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr19_-_39390168
|
43.999
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr16_+_58498725
|
39.002
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58534045
|
35.842
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr5_-_149535420
|
33.039
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr16_+_58497993
|
32.952
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr1_+_151254740
|
32.639
|
NM_020832
|
ZNF687
|
zinc finger protein 687
|
|
chr16_+_58497548
|
31.350
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr17_+_7554239
|
31.183
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr16_+_11762235
|
30.404
|
NM_003498
|
SNN
|
stannin
|
|
chr2_+_242641310
|
29.450
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr2_+_203499819
|
28.772
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr11_-_61684981
|
27.773
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr7_+_138916230
|
26.448
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr1_+_46669005
|
26.130
|
NM_001013615
|
C1orf190
|
chromosome 1 open reading frame 190
|
|
chr17_-_7197866
|
25.991
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr7_-_17980102
|
25.799
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr11_-_59383616
|
24.621
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr1_+_204913353
|
24.593
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr1_-_200992824
|
24.290
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr6_-_109330190
|
23.386
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr12_+_4918341
|
22.852
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr17_-_28257017
|
22.788
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr6_-_44280986
|
22.639
|
NM_020745
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr4_+_95679075
|
21.509
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr9_-_130829598
|
20.934
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr17_-_37764165
|
20.586
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr2_-_86564632
|
20.296
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr19_-_18632886
|
20.250
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr19_+_41284068
|
20.197
|
NM_016154
|
RAB4B
|
RAB4B, member RAS oncogene family
|
|
chr19_+_57752051
|
19.807
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr6_-_109330752
|
19.468
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr19_+_50015535
|
19.408
|
NM_001136019
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr11_-_2906960
|
19.354
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr5_-_149792370
|
19.266
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr6_-_39902140
|
19.257
|
NM_001075098
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr3_-_39321929
|
18.866
|
NM_001171172
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr12_-_49504646
|
18.648
|
NM_018113
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr11_-_119234859
|
18.627
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_43864433
|
18.487
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr19_+_50016435
|
18.464
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr2_-_86565205
|
18.459
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr14_+_24583979
|
18.389
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_109415514
|
18.357
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chrX_-_107019001
|
18.346
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_+_18218569
|
18.312
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr7_-_44365019
|
17.978
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr12_-_123451031
|
17.940
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr1_+_204797774
|
17.795
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr5_+_150399980
|
17.773
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr3_-_39321479
|
17.476
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr14_+_24583905
|
17.429
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr12_-_49392909
|
17.346
|
NM_015086
|
DDN
|
dendrin
|
|
chr9_-_77502262
|
17.090
|
NM_001177311
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr5_-_140998567
|
17.089
|
NM_001079812
NM_005219
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr11_-_117748200
|
16.777
|
NM_001164831
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr19_+_18208596
|
16.754
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr17_-_38256905
|
16.643
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr12_+_7023490
|
16.627
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr1_-_46598250
|
16.561
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr4_+_1795004
|
16.520
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr1_+_156119734
|
16.408
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_-_36235422
|
16.046
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr8_-_10587942
|
15.959
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr10_+_111985628
|
15.824
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chrX_-_106960268
|
15.751
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_101702304
|
15.729
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_+_27561006
|
15.688
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr6_-_33267128
|
15.586
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr9_-_10612722
|
15.495
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr5_+_65017993
|
15.485
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr8_+_26240522
|
15.471
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr2_-_73298815
|
15.466
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr3_+_49591897
|
15.363
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chrX_-_49056638
|
15.313
|
NM_003179
|
SYP
|
synaptophysin
|
|
chr8_-_103136486
|
15.309
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr9_-_101470836
|
15.299
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr5_+_137774632
|
15.256
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr10_+_111967302
|
15.248
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr9_+_37753769
|
15.131
|
NM_144964
|
RG9MTD3
|
RNA (guanine-9-) methyltransferase domain containing 3
|
|
chr1_-_11866079
|
14.936
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr1_+_156123321
|
14.867
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr9_-_72287097
|
14.770
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr6_+_37137882
|
14.647
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr17_-_8055684
|
14.639
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr3_-_39322763
|
14.518
|
NM_001171171
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr19_+_19322763
|
14.368
|
NM_004386
|
NCAN
|
neurocan
|
|
chr1_-_203155910
|
14.320
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr3_-_66024508
|
14.192
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr6_-_39895317
|
14.190
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr14_+_61788398
|
14.135
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr3_-_39323183
|
14.082
|
NM_001171174
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr15_+_41851153
|
14.043
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr9_-_127269689
|
13.938
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr16_+_67927170
|
13.934
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr10_-_103454687
|
13.850
|
NM_022039
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr2_-_62081049
|
13.818
|
NM_001201543
NM_032180
|
FAM161A
|
family with sequence similarity 161, member A
|
|
chr5_-_115910406
|
13.775
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr2_-_175351755
|
13.767
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr11_-_1593149
|
13.745
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr7_-_97881436
|
13.712
|
NM_015395
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chr8_-_67579446
|
13.697
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr10_-_62149487
|
13.693
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_+_111969988
|
13.620
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr19_+_40973049
|
13.612
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr4_+_7045142
|
13.571
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr16_-_4292052
|
13.492
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr15_+_65134081
|
13.417
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr17_-_78450247
|
13.405
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chrX_-_106959597
|
13.358
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr22_-_51021337
|
13.329
|
NM_005198
|
CHKB
CHKB-CPT1B
|
choline kinase beta
CHKB-CPT1B readthrough (non-protein coding)
|
|
chr17_-_10101854
|
13.299
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr9_+_91933384
|
13.296
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr5_-_149492934
|
13.285
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr9_-_77503009
|
13.132
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr3_+_9691116
|
13.005
|
NM_001077525
NM_001077526
NM_022485
|
MTMR14
|
myotubularin related protein 14
|
|
chr11_-_62599518
|
12.973
|
NM_001244666
NM_003164
|
STX5
|
syntaxin 5
|
|
chr16_+_57769626
|
12.890
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr13_+_24153426
|
12.847
|
NM_001204458
NM_018647
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr20_-_4804067
|
12.782
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr8_-_102803438
|
12.742
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr5_-_131826426
|
12.716
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr12_-_51477299
|
12.592
|
NM_030809
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr20_-_33680601
|
12.591
|
NM_015638
NM_199368
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr1_+_35225341
|
12.542
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr6_+_36210944
|
12.439
|
NM_173676
NM_001145716
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr14_+_92790151
|
12.373
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr9_-_131534186
|
12.363
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr3_+_47324329
|
12.363
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr15_+_75287829
|
12.343
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr8_-_121824287
|
12.267
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr11_+_45868956
|
12.242
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr11_-_77899640
|
12.072
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr19_-_14117096
|
12.041
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr5_-_137071703
|
11.905
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr12_-_53625834
|
11.902
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr17_-_27916602
|
11.861
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr8_-_103137134
|
11.720
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr15_+_42066631
|
11.687
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr17_+_15602890
|
11.630
|
NM_001130842
NM_020652
|
ZNF286A
|
zinc finger protein 286A
|
|
chr3_-_114343791
|
11.591
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_+_45868668
|
11.440
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr5_-_132948222
|
11.367
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr1_-_151118932
|
11.335
|
NM_001178061
NM_001178062
NM_030913
|
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C
|
|
chr22_-_19279183
|
11.193
|
NM_001835
NM_007098
|
CLTCL1
|
clathrin, heavy chain-like 1
|
|
chr2_-_37311458
|
11.125
|
NM_019024
|
HEATR5B
|
HEAT repeat containing 5B
|
|
chr2_-_200820458
|
11.096
|
NM_001039693
|
TYW5
|
tRNA-yW synthesizing protein 5
|
|
chr11_+_45907046
|
11.040
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr14_+_92789536
|
10.998
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr20_-_4795768
|
10.995
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr2_+_218990735
|
10.956
|
NM_001557
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr17_-_9862765
|
10.949
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr20_+_62289162
|
10.887
|
NM_016434
NM_032957
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr2_-_172967234
|
10.871
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr4_-_8160435
|
10.835
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr1_-_6240082
|
10.816
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr2_-_97304080
|
10.779
|
NM_001115016
NM_017991
|
KANSL3
|
KAT8 regulatory NSL complex subunit 3
|
|
chr12_+_9067130
|
10.765
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr17_-_72889704
|
10.721
|
NM_178128
|
FADS6
|
fatty acid desaturase domain family, member 6
|
|
chr3_-_50649110
|
10.643
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr1_+_11866206
|
10.621
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr10_-_62493282
|
10.610
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrX_+_57618268
|
10.593
|
NM_007157
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr20_+_54933982
|
10.588
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr13_-_28543290
|
10.582
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr1_+_118148506
|
10.553
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr3_+_133464883
|
10.526
|
NM_001063
|
TF
|
transferrin
|
|
chr17_-_56406120
|
10.525
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr10_+_101419169
|
10.521
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr9_-_98079856
|
10.495
|
NM_000136
NM_001243744
|
FANCC
|
Fanconi anemia, complementation group C
|
|
chr12_+_123319977
|
10.476
|
NM_003959
|
HIP1R
|
huntingtin interacting protein 1 related
|
|
chr19_-_47734215
|
10.427
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr15_-_64648359
|
10.413
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr1_-_156786500
|
10.116
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr12_+_113229548
|
10.111
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr11_-_66115016
|
10.042
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr2_-_217560247
|
9.957
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr6_+_36238236
|
9.953
|
NM_001145717
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr5_+_139493689
|
9.940
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr12_+_117176085
|
9.930
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr17_+_72744746
|
9.915
|
NM_004252
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chrX_-_18372777
|
9.892
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr17_+_34431219
|
9.847
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr2_-_69870976
|
9.836
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr10_-_74114865
|
9.821
|
NM_001002762
NM_017626
|
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12
|
|
chr6_+_111580481
|
9.801
|
NM_153369
|
KIAA1919
|
KIAA1919
|
|
chr22_+_38004454
|
9.785
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr6_+_42981840
|
9.774
|
NM_057161
|
KLHDC3
|
kelch domain containing 3
|
|
chr3_-_120068077
|
9.642
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr9_-_77502635
|
9.568
|
NM_001177310
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr15_+_89346663
|
9.528
|
NM_001135
NM_013227
|
ACAN
|
aggrecan
|
|
chrX_+_122318095
|
9.508
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr2_-_182545212
|
9.487
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr11_-_102962903
|
9.471
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr14_-_75593720
|
9.400
|
NM_033116
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr6_+_106546736
|
9.371
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_43543877
|
9.257
|
NM_006502
|
POLH
|
polymerase (DNA directed), eta
|
|
chr1_+_11751778
|
9.238
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr5_-_112630557
|
9.232
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|