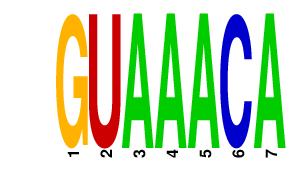
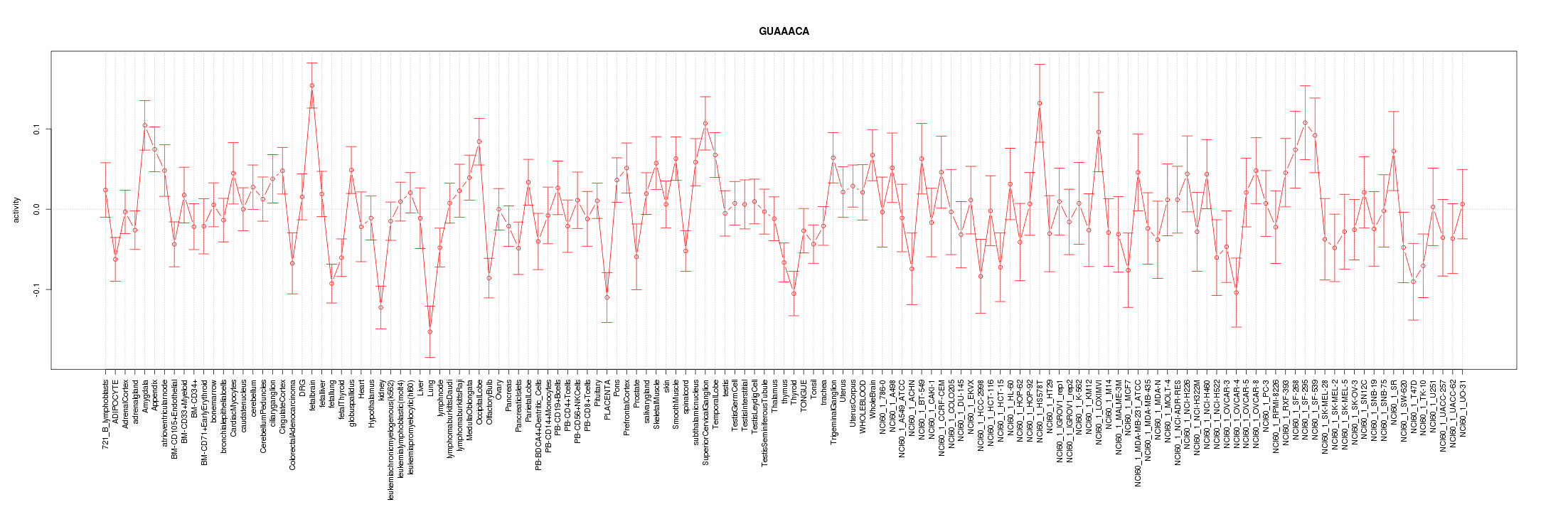
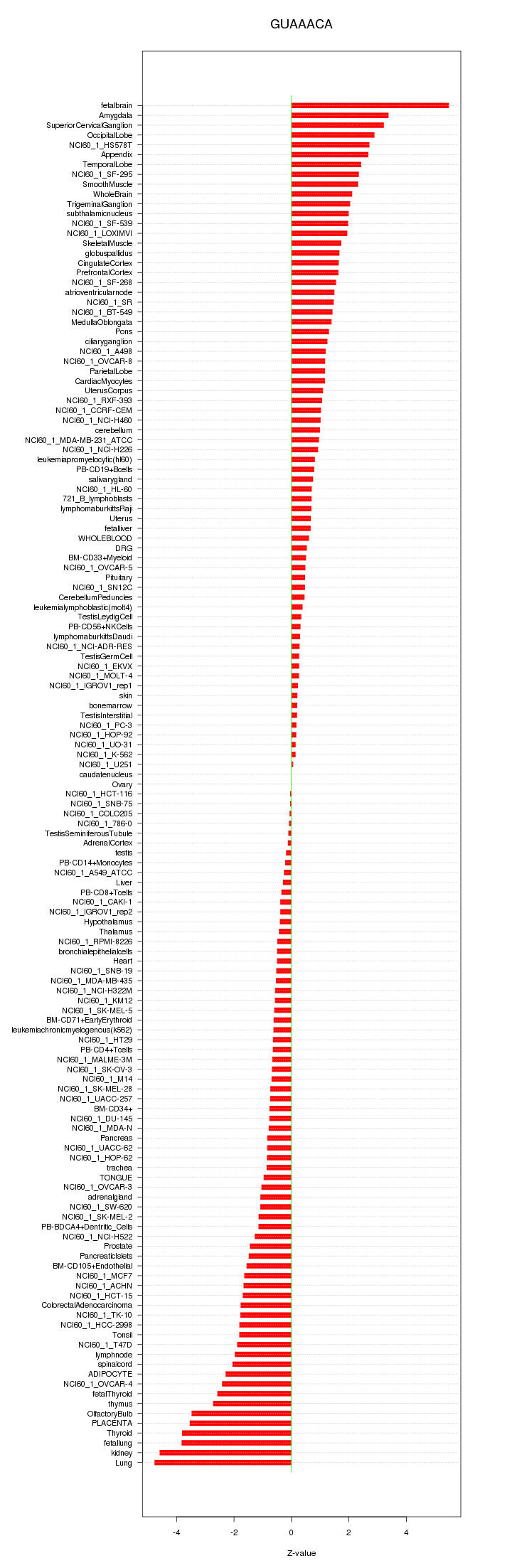
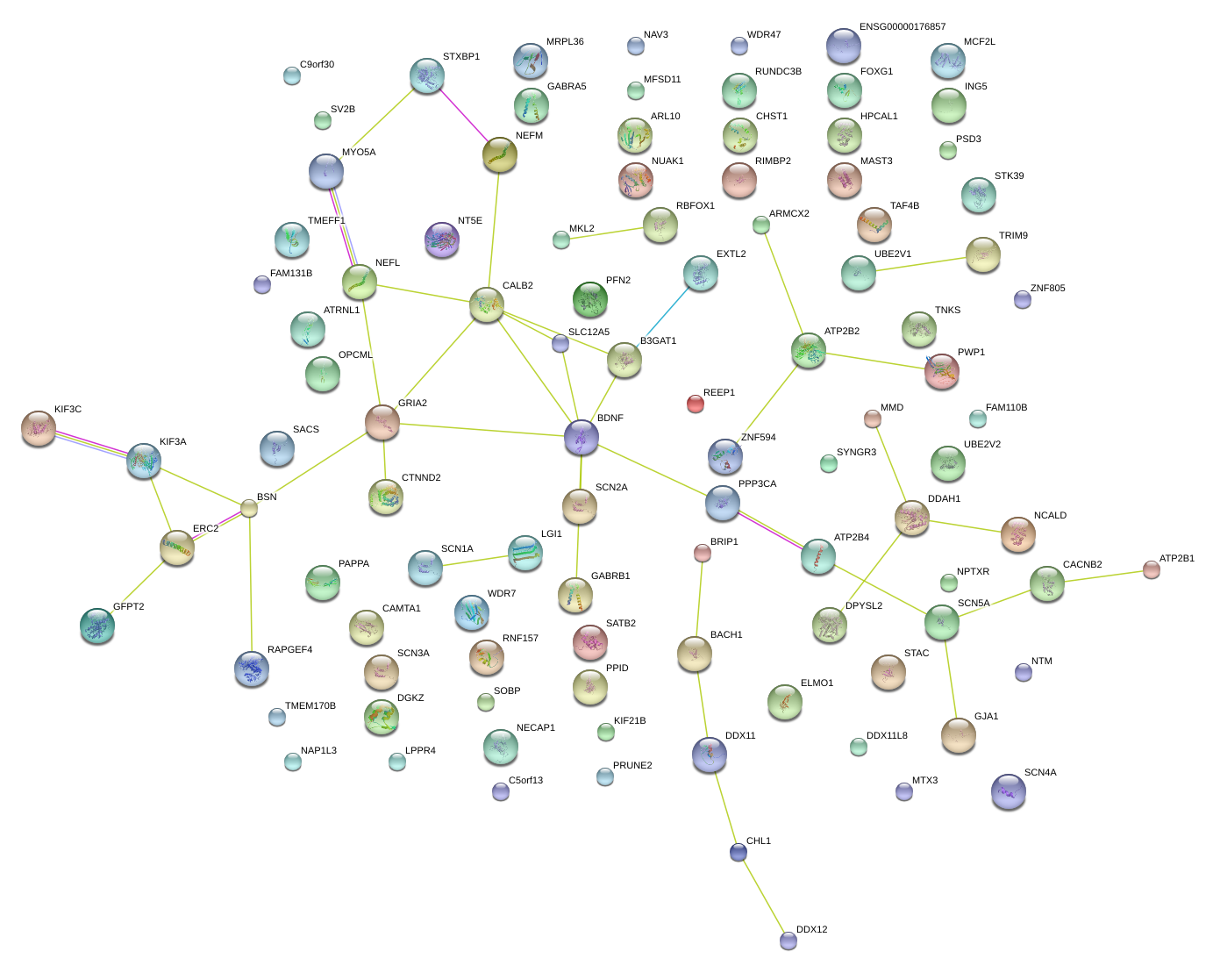
|
chr8_-_24814028
|
13.975
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr16_+_2039945
|
12.205
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr4_+_175204827
|
11.303
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr20_+_44650260
|
11.296
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr4_-_102268626
|
11.058
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr8_+_24772454
|
10.310
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_24771268
|
9.821
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr2_-_86565205
|
9.759
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr20_+_44657812
|
9.744
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr8_-_18871163
|
9.288
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_-_86564632
|
9.056
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr3_+_238274
|
8.720
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr8_+_26371461
|
8.604
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_-_106533810
|
8.545
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr4_+_158141735
|
8.483
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr4_+_47033294
|
8.406
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr19_+_18208596
|
8.332
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr6_+_107811272
|
7.910
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr12_+_78225068
|
7.758
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chrX_-_92928566
|
7.627
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr8_-_103136486
|
7.534
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr8_-_103137134
|
7.371
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr16_+_6069094
|
7.345
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr9_+_130374466
|
7.342
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr9_+_103235489
|
7.337
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr19_+_57752051
|
7.209
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr8_+_26435339
|
7.166
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_+_8234796
|
7.146
|
NM_015509
|
NECAP1
|
NECAP endocytosis associated 1
|
|
chr8_-_18666295
|
6.786
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr7_-_143059144
|
6.438
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr16_+_6823809
|
6.414
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr3_+_49591897
|
6.369
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr7_-_143059696
|
6.333
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr11_-_27743604
|
6.240
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721975
|
6.217
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722446
|
6.181
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_131002409
|
6.121
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr11_-_27722599
|
6.012
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_11903964
|
5.956
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr13_-_24007822
|
5.896
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr11_-_134281725
|
5.891
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr7_-_37393271
|
5.855
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr9_+_103204187
|
5.852
|
NM_001198812
|
C9orf30-TMEFF1
C9orf30
|
C9orf30-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr11_-_27681195
|
5.849
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_99729847
|
5.840
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr12_-_90049818
|
5.806
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr16_+_14164879
|
5.760
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr2_-_166060552
|
5.693
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr1_-_101360405
|
5.643
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr5_-_111093792
|
5.603
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_36421944
|
5.577
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr18_+_54318561
|
5.518
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr6_+_121756744
|
5.511
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr11_-_45687135
|
5.353
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr11_-_27721179
|
5.307
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_-_102803438
|
5.268
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr10_+_18429605
|
5.204
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr6_+_11538486
|
5.189
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr9_-_79520878
|
5.138
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr13_+_113623496
|
5.079
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr10_+_116853118
|
4.997
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr16_+_7382750
|
4.964
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr2_-_169103886
|
4.937
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr3_-_56502364
|
4.887
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr5_-_132073143
|
4.885
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr5_-_111312522
|
4.821
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_74236272
|
4.795
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr5_-_79286793
|
4.790
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr1_-_200992824
|
4.751
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr5_-_111093251
|
4.714
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_85930733
|
4.700
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_-_111093030
|
4.681
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_26205437
|
4.678
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr11_-_27723061
|
4.630
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_179780314
|
4.618
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr3_-_149688638
|
4.599
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr14_+_29236171
|
4.493
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr17_-_53499055
|
4.389
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr7_+_87257704
|
4.327
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr8_+_48920969
|
4.298
|
NM_003350
|
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr10_+_18689512
|
4.248
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr15_+_27111865
|
4.240
|
NM_000810
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr10_+_95517565
|
4.207
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr5_+_175792444
|
4.196
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr6_+_86159301
|
4.150
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr14_-_51562421
|
4.146
|
NM_015163
NM_052978
|
TRIM9
|
tripartite motif containing 9
|
|
chr7_-_37488894
|
4.078
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr22_-_39240016
|
4.071
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr7_-_37488554
|
4.026
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr11_+_131240370
|
3.989
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr15_+_27112106
|
3.953
|
NM_001165037
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr18_+_23806385
|
3.944
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr8_+_9413439
|
3.913
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr15_-_52821004
|
3.903
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr7_-_37024664
|
3.879
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr11_-_27741192
|
3.870
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_+_46368881
|
3.866
|
NM_001199266
NM_001199267
NM_001199268
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr16_+_71392615
|
3.796
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr21_+_30671217
|
3.773
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr3_-_10547267
|
3.768
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_-_111092918
|
3.757
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_173600519
|
3.730
|
NM_007023
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr1_-_109584589
|
3.718
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr17_+_74733468
|
3.698
|
NM_001242532
NM_001242533
NM_001242535
NM_001242536
NM_001242537
NM_024311
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr10_+_18549583
|
3.659
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr2_+_242641310
|
3.605
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr8_+_58906968
|
3.587
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr11_+_46366816
|
3.551
|
NM_201533
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr2_-_200335988
|
3.528
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr15_+_91643429
|
3.522
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_+_6845381
|
3.520
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr2_-_166930130
|
3.519
|
NM_001165963
NM_001165964
NM_006920
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr4_-_159644439
|
3.514
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr2_+_166152282
|
3.513
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr15_-_34502269
|
3.512
|
NM_024713
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr4_+_41362750
|
3.499
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_-_34316325
|
3.498
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr6_-_169654047
|
3.493
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr5_-_142783225
|
3.489
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chrX_+_117480027
|
3.468
|
NM_001184965
NM_001184966
NM_019045
|
WDR44
|
WD repeat domain 44
|
|
chr7_+_35840595
|
3.423
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr2_+_136288953
|
3.419
|
NM_015361
|
R3HDM1
|
R3H domain containing 1
|
|
chr5_-_121414011
|
3.365
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr2_+_166150340
|
3.363
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr17_-_36956145
|
3.329
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr9_-_138987096
|
3.326
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr1_+_244214552
|
3.313
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr10_+_12171580
|
3.286
|
NM_001142627
NM_001142628
NM_018144
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr2_+_173686314
|
3.262
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr3_-_9291251
|
3.251
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr15_+_91643181
|
3.222
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_-_84464728
|
3.220
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr4_-_36245893
|
3.180
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr1_-_86043932
|
3.172
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_16847070
|
3.157
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr2_-_167005641
|
3.150
|
NM_001202435
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr7_-_79082870
|
3.149
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr16_+_66914380
|
3.118
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr7_+_29846112
|
3.107
|
NM_001080529
|
WIPF3
|
WAS/WASL interacting protein family, member 3
|
|
chr6_-_128841432
|
3.106
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr5_-_121412805
|
3.084
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr10_+_18629613
|
3.057
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr13_+_113656027
|
3.053
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr10_-_75255747
|
3.052
|
NM_001142353
NM_001142354
NM_021132
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme
|
|
chr5_-_72744351
|
3.028
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr11_-_27742224
|
3.023
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_118846029
|
3.018
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr1_+_16062808
|
2.972
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr8_+_99129488
|
2.963
|
NM_001145860
NM_001145861
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr3_-_50605062
|
2.957
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr8_+_26240522
|
2.950
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr5_-_142815076
|
2.945
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr3_-_185216545
|
2.914
|
NM_080652
|
TMEM41A
|
transmembrane protein 41A
|
|
chr1_-_85155885
|
2.886
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr3_+_15469052
|
2.885
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr5_-_53606387
|
2.878
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr5_-_114598547
|
2.862
|
NM_005023
|
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr15_-_37393364
|
2.848
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_200322699
|
2.826
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr17_-_74449272
|
2.815
|
NM_022066
|
UBE2O
|
ubiquitin-conjugating enzyme E2O
|
|
chr6_+_158733691
|
2.799
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr19_-_4559713
|
2.796
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr8_+_132916339
|
2.771
|
NM_015137
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr5_-_111091947
|
2.766
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_-_33146543
|
2.764
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr11_-_88796815
|
2.737
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr5_+_150827138
|
2.720
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr11_+_46354454
|
2.705
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr8_+_99130067
|
2.696
|
NM_015029
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr4_+_41614911
|
2.691
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr19_+_32896515
|
2.680
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr8_+_107282343
|
2.658
|
NM_001198533
NM_018002
|
OXR1
|
oxidation resistance 1
|
|
chr8_+_107669895
|
2.649
|
NM_181354
|
OXR1
|
oxidation resistance 1
|
|
chr13_-_50367027
|
2.649
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr11_+_112831968
|
2.626
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_-_76953561
|
2.624
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr9_-_23821842
|
2.616
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr14_-_65438794
|
2.605
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr5_+_138609440
|
2.602
|
NM_001194954
NM_199189
|
MATR3
|
matrin 3
|
|
chr3_+_123813527
|
2.597
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr12_-_121734543
|
2.595
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr7_-_35077652
|
2.579
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr7_+_18126543
|
2.566
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr10_-_23003443
|
2.565
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr2_-_200329810
|
2.560
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr22_+_26565424
|
2.556
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr14_+_79745653
|
2.503
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr2_-_235405692
|
2.497
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_+_17942584
|
2.490
|
NM_024052
|
C17orf39
|
chromosome 17 open reading frame 39
|
|
chr7_+_18548899
|
2.489
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr16_+_82660390
|
2.480
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr12_-_42631982
|
2.469
|
NM_001190977
|
YAF2
|
YY1 associated factor 2
|
|
chr1_-_151688791
|
2.465
|
NM_001172648
NM_001172649
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr3_-_101395611
|
2.464
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr4_+_4387982
|
2.446
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr20_+_56884749
|
2.443
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr7_+_102553343
|
2.431
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr9_-_23821477
|
2.422
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_113542970
|
2.420
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr13_-_22033505
|
2.385
|
NM_153251
|
ZDHHC20
|
zinc finger, DHHC-type containing 20
|