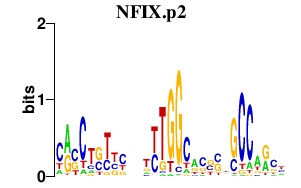
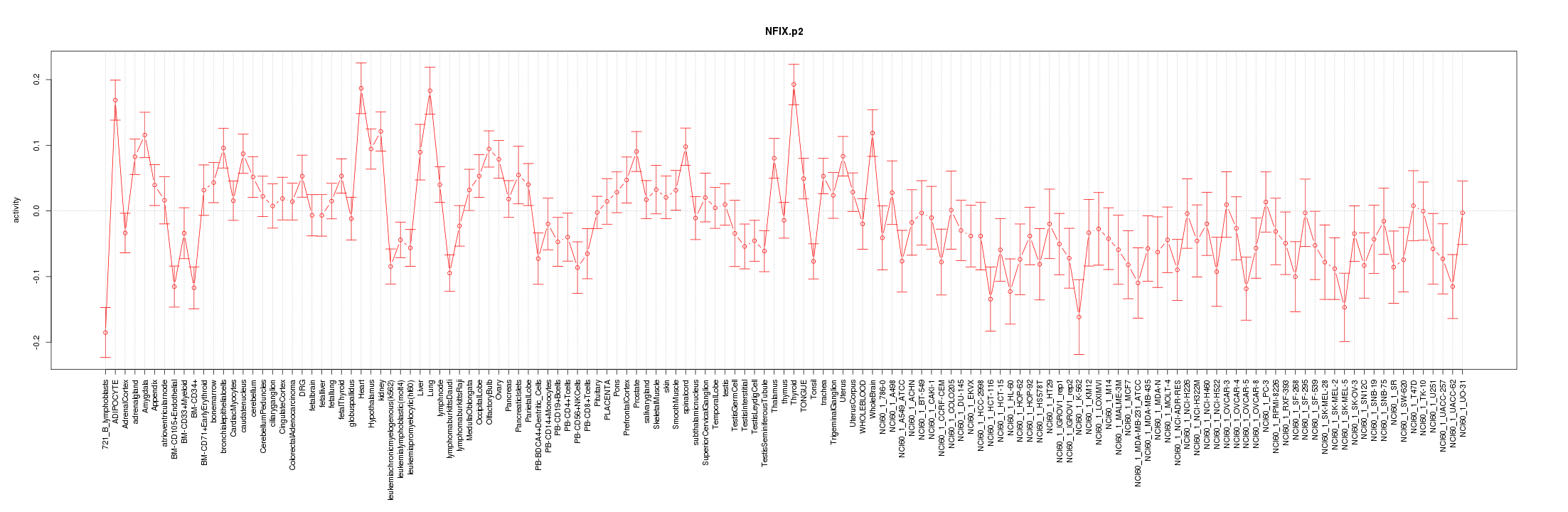
|
chr5_+_150399980
|
38.927
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr5_+_150400132
|
38.038
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr10_+_124220990
|
22.255
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr15_-_72668184
|
19.994
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr9_+_124030370
|
19.182
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr9_+_124030455
|
18.890
|
|
GSN
|
gelsolin
|
|
chr11_-_72353435
|
18.288
|
NM_001143839
NM_001243784
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr11_-_46722160
|
17.311
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr12_+_26348268
|
16.790
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr18_-_74691951
|
16.206
|
|
MBP
|
myelin basic protein
|
|
chr11_-_46722139
|
16.099
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr9_+_124030413
|
16.059
|
|
GSN
|
gelsolin
|
|
chr17_+_37844325
|
16.012
|
NM_001005862
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr11_-_61735108
|
14.822
|
NM_002032
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr17_-_37844266
|
14.793
|
NM_033419
|
PGAP3
|
post-GPI attachment to proteins 3
|
|
chr15_+_91769329
|
14.565
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr10_+_88728187
|
14.263
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr20_-_44539542
|
13.805
|
|
PLTP
|
phospholipid transfer protein
|
|
chr1_-_36235422
|
13.693
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr11_-_46722115
|
13.682
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr1_-_2322996
|
13.271
|
|
MORN1
|
MORN repeat containing 1
|
|
chr1_+_44584521
|
13.097
|
NM_173484
|
KLF17
|
Kruppel-like factor 17
|
|
chrX_+_13671224
|
12.887
|
NM_152634
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr16_-_4852571
|
12.782
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr8_+_24771268
|
12.395
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr3_-_50396892
|
11.949
|
|
TMEM115
|
transmembrane protein 115
|
|
chr1_+_1260289
|
11.893
|
|
|
|
|
chr11_-_57004674
|
11.753
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr1_-_2322990
|
11.568
|
NM_024848
|
MORN1
|
MORN repeat containing 1
|
|
chr9_+_35605339
|
11.450
|
|
TESK1
|
testis-specific kinase 1
|
|
chr8_+_24771276
|
11.283
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr3_-_28390506
|
11.280
|
NM_001134432
NM_001134433
NM_022461
|
AZI2
|
5-azacytidine induced 2
|
|
chr11_-_61735067
|
11.204
|
|
FTH1
CPSF7
|
ferritin, heavy polypeptide 1
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chr9_+_35605275
|
10.993
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr17_-_28562953
|
10.978
|
NM_001045
|
SLC6A4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4
|
|
chr17_-_79105747
|
10.975
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr7_+_108210291
|
10.811
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr17_-_80656461
|
10.694
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr11_-_12030128
|
10.418
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_61734896
|
10.373
|
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr7_+_108210308
|
10.363
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr13_+_48807307
|
10.277
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr9_+_130911731
|
10.243
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr3_-_50396930
|
10.132
|
NM_007024
|
TMEM115
|
transmembrane protein 115
|
|
chr3_+_130745693
|
10.076
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr1_-_156217717
|
10.057
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr11_-_125366048
|
9.774
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr3_-_42003460
|
9.632
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr2_+_39664544
|
9.562
|
|
LOC728730
|
uncharacterized LOC728730
|
|
chr3_-_28390280
|
9.536
|
|
AZI2
|
5-azacytidine induced 2
|
|
chr9_-_94124149
|
9.486
|
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr19_-_11373117
|
9.369
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr10_-_100206643
|
9.300
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr17_+_18128868
|
9.297
|
NM_004140
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila)
|
|
chr9_+_34179058
|
9.275
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr6_+_41040713
|
9.262
|
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr19_-_3028901
|
9.245
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr3_+_138067678
|
9.228
|
NM_001252092
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr12_-_45270071
|
9.075
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr7_+_108210188
|
9.069
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr3_+_133495925
|
9.056
|
|
TF
|
transferrin
|
|
chr10_-_7661613
|
9.037
|
NM_032817
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr9_-_130616592
|
8.949
|
|
ENG
|
endoglin
|
|
chr16_+_29985185
|
8.897
|
NM_001252043
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr12_+_51985016
|
8.837
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr15_+_74466887
|
8.832
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr6_+_30851860
|
8.721
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_-_109330190
|
8.702
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chrX_+_21857786
|
8.634
|
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr7_+_156742402
|
8.574
|
NM_138400
|
NOM1
|
nucleolar protein with MIF4G domain 1
|
|
chr3_+_138067969
|
8.514
|
NM_001252090
|
MRAS
|
muscle RAS oncogene homolog
|
|
chrX_+_21857775
|
8.506
|
|
|
|
|
chr9_+_34179036
|
8.493
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr11_-_117166247
|
8.452
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr8_-_11725509
|
8.280
|
|
CTSB
|
cathepsin B
|
|
chr3_+_138067444
|
8.245
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr11_-_12030539
|
8.221
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr9_+_34179002
|
8.205
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr6_-_46459010
|
8.090
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_+_30156326
|
7.973
|
NM_024310
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr2_+_241375030
|
7.958
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr3_+_35681008
|
7.907
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_+_53443834
|
7.905
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr4_-_99579540
|
7.821
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr19_-_6720585
|
7.811
|
NM_000064
|
C3
|
complement component 3
|
|
chr5_+_23507723
|
7.798
|
NM_020227
|
PRDM9
|
PR domain containing 9
|
|
chrX_+_21857745
|
7.711
|
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr9_+_34179023
|
7.625
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr9_+_115913227
|
7.512
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr8_-_33424642
|
7.475
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr4_+_3294711
|
7.459
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_+_115397454
|
7.332
|
NM_003176
|
SYCP1
|
synaptonemal complex protein 1
|
|
chr11_+_64851691
|
7.327
|
NM_006782
|
ZFPL1
|
zinc finger protein-like 1
|
|
chr16_+_56703725
|
7.049
|
NM_005951
|
MT1H
|
metallothionein 1H
|
|
chr22_-_36019363
|
7.037
|
NM_203377
|
MB
|
myoglobin
|
|
chr12_+_108909049
|
6.994
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr10_+_73079044
|
6.881
|
|
SLC29A3
|
solute carrier family 29 (nucleoside transporters), member 3
|
|
chr9_-_113800243
|
6.798
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr13_+_27131879
|
6.794
|
|
WASF3
|
WAS protein family, member 3
|
|
chr10_+_38299525
|
6.742
|
NM_006954
NM_006974
|
ZNF33A
|
zinc finger protein 33A
|
|
chr10_-_105992062
|
6.686
|
NM_025145
|
WDR96
|
WD repeat domain 96
|
|
chr9_-_130616760
|
6.664
|
|
ENG
|
endoglin
|
|
chr13_+_48807492
|
6.579
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr8_-_7212868
|
6.539
|
|
FAM66B
|
family with sequence similarity 66, member B
|
|
chr16_+_29985213
|
6.528
|
|
TAOK2
|
TAO kinase 2
|
|
chr4_-_85419386
|
6.447
|
NM_006168
|
NKX6-1
|
NK6 homeobox 1
|
|
chr9_+_133320093
|
6.445
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr1_+_77333121
|
6.443
|
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr8_-_11725587
|
6.432
|
|
CTSB
|
cathepsin B
|
|
chr21_-_48024985
|
6.418
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr11_-_12030622
|
6.407
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr19_+_45312294
|
6.350
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr22_+_19701968
|
6.269
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr12_+_50355278
|
6.249
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr6_-_109330752
|
6.196
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chrX_+_21857655
|
6.194
|
NM_015884
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr15_+_67358161
|
6.175
|
|
SMAD3
|
SMAD family member 3
|
|
chr22_-_36903033
|
6.166
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chrX_+_10124984
|
6.111
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr22_-_44258210
|
6.062
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr11_-_64527369
|
6.039
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr10_+_73079009
|
6.027
|
NM_001174098
NM_018344
|
SLC29A3
|
solute carrier family 29 (nucleoside transporters), member 3
|
|
chr2_+_3642421
|
5.993
|
NM_024027
NM_199235
|
COLEC11
|
collectin sub-family member 11
|
|
chr4_-_8442427
|
5.972
|
NM_001101667
NM_003501
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr13_-_77460432
|
5.969
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr9_+_133320247
|
5.957
|
|
ASS1
|
argininosuccinate synthase 1
|
|
chr2_+_149632772
|
5.937
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr2_-_232395108
|
5.917
|
NM_006056
|
NMUR1
|
neuromedin U receptor 1
|
|
chr6_+_41040687
|
5.910
|
NM_002505
NM_021705
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr5_-_1634099
|
5.865
|
|
LOC728613
|
programmed cell death 6 pseudogene
|
|
chr13_+_48807250
|
5.841
|
NM_021999
|
ITM2B
|
integral membrane protein 2B
|
|
chrX_-_102319096
|
5.747
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr5_+_141303463
|
5.745
|
|
KIAA0141
|
KIAA0141
|
|
chr16_+_1290677
|
5.715
|
NM_003294
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr9_-_94124194
|
5.663
|
NM_001698
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr11_-_64528186
|
5.655
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr4_-_73935408
|
5.647
|
NM_173827
|
COX18
|
COX18 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr4_+_1795004
|
5.572
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr19_+_38893828
|
5.535
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr16_-_30905262
|
5.499
|
|
BCL7C
|
B-cell CLL/lymphoma 7C
|
|
chr3_+_44596596
|
5.485
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr11_-_49229878
|
5.459
|
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1
|
|
chr1_+_46859918
|
5.410
|
NM_001441
|
FAAH
|
fatty acid amide hydrolase
|
|
chr2_-_223163333
|
5.363
|
NM_000438
NM_001127366
NM_013942
NM_181457
NM_181458
NM_181459
NM_181460
NM_181461
|
PAX3
|
paired box 3
|
|
chr12_+_26348488
|
5.361
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr1_-_236228476
|
5.359
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr19_+_46195898
|
5.330
|
|
QPCTL
|
glutaminyl-peptide cyclotransferase-like
|
|
chr8_+_11142314
|
5.316
|
|
MTMR9
|
myotubularin related protein 9
|
|
chr19_+_859658
|
5.308
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr12_-_45269953
|
5.307
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_160822625
|
5.282
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr8_+_75262616
|
5.229
|
NM_018972
NM_001040875
|
GDAP1
|
ganglioside-induced differentiation-associated protein 1
|
|
chr4_-_109684053
|
5.228
|
NM_001146590
NM_001146627
NM_031279
|
AGXT2L1
|
alanine-glyoxylate aminotransferase 2-like 1
|
|
chr16_+_66968390
|
5.227
|
|
CES2
|
carboxylesterase 2
|
|
chr1_+_77333158
|
5.198
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr9_-_130616957
|
5.194
|
|
ENG
|
endoglin
|
|
chr2_+_149633173
|
5.194
|
|
|
|
|
chr2_-_56151273
|
5.075
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr16_-_89556886
|
5.070
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr6_-_109761707
|
5.063
|
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr17_-_34890610
|
5.043
|
|
MYO19
|
myosin XIX
|
|
chr2_+_149632836
|
5.043
|
|
KIF5C
|
kinesin family member 5C
|
|
chr17_+_32597239
|
5.024
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr19_+_57791794
|
4.999
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr5_-_157002705
|
4.995
|
|
|
|
|
chr20_+_43344005
|
4.977
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr3_-_46506342
|
4.977
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr15_-_71184611
|
4.972
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr9_+_104161118
|
4.970
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr4_+_156680125
|
4.956
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr1_-_13390747
|
4.915
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr17_+_18012019
|
4.903
|
NM_016239
|
MYO15A
|
myosin XVA
|
|
chr8_-_11725622
|
4.895
|
|
CTSB
|
cathepsin B
|
|
chr15_-_72668301
|
4.894
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr5_+_141303366
|
4.870
|
NM_001142603
NM_014773
|
KIAA0141
|
KIAA0141
|
|
chr19_+_38755097
|
4.820
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr14_-_93582143
|
4.809
|
NM_001142593
NM_001142594
NM_014216
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase
|
|
chr8_-_41166931
|
4.806
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr2_-_71221951
|
4.780
|
|
TEX261
|
testis expressed 261
|
|
chr16_-_30546145
|
4.759
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr7_-_150038689
|
4.748
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr3_+_10290204
|
4.725
|
|
TATDN2
|
TatD DNase domain containing 2
|
|
chr5_+_140797270
|
4.693
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr15_-_26874236
|
4.652
|
NM_001191321
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chrX_-_102319080
|
4.645
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr3_-_16646244
|
4.644
|
NM_001190811
|
DAZL
|
deleted in azoospermia-like
|
|
chr1_-_182360889
|
4.631
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr8_-_54755821
|
4.616
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr14_-_93651182
|
4.614
|
|
MOAP1
|
modulator of apoptosis 1
|
|
chr1_+_244816434
|
4.591
|
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr13_+_28494136
|
4.564
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr13_-_20357081
|
4.559
|
|
PSPC1
|
paraspeckle component 1
|
|
chr20_-_32262168
|
4.545
|
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr14_-_106208695
|
4.537
|
|
|
|
|
chr1_+_10003485
|
4.537
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr6_+_31795395
|
4.516
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr3_+_184279539
|
4.515
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr4_-_120548329
|
4.478
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr12_+_72666462
|
4.431
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr10_+_11784355
|
4.401
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|