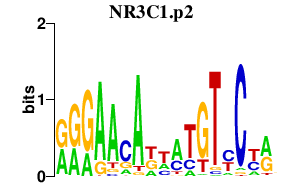
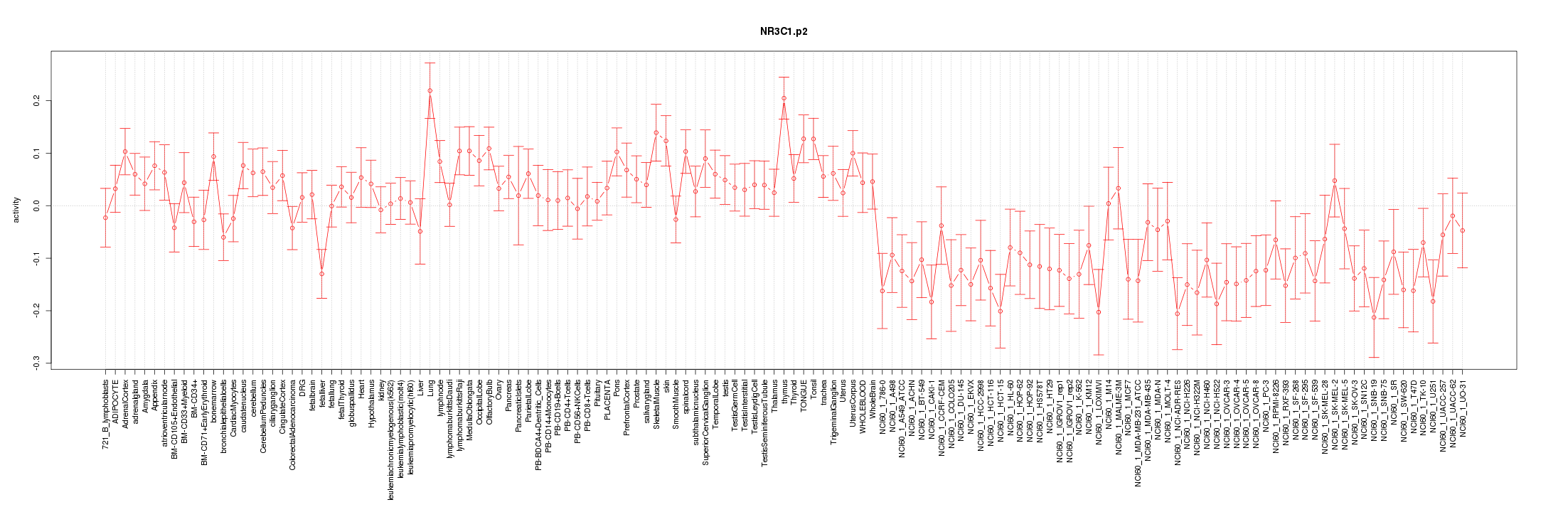
|
chr12_-_91572328
|
24.933
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr21_-_46340776
|
22.777
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr11_+_73358593
|
22.081
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr21_-_46340821
|
22.046
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr7_+_23286390
|
21.590
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr22_+_22712091
|
20.863
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr7_+_23286386
|
20.799
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286384
|
20.616
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr3_-_187463227
|
20.574
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_237172987
|
19.131
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr7_+_20655244
|
18.844
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr19_-_36643770
|
18.811
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr10_+_70847817
|
17.450
|
NM_002727
|
SRGN
|
serglycin
|
|
chr7_-_124405029
|
15.140
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr8_-_110655418
|
14.435
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr17_-_8055684
|
13.099
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr6_-_6320875
|
12.916
|
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr1_+_44115804
|
12.884
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chrY_+_16168097
|
12.744
|
NM_181880
NM_004679
|
VCY1B
VCX3A
VCY
|
variable charge, Y-linked 1B
variable charge, X-linked 3A
variable charge, Y-linked
|
|
chr19_+_15783827
|
12.527
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr1_-_153085963
|
12.189
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr6_-_6320897
|
12.130
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr18_-_53068755
|
11.781
|
|
TCF4
|
transcription factor 4
|
|
chr16_-_68482373
|
11.739
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr6_-_32191779
|
11.550
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr1_-_204459445
|
11.450
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr7_-_37393271
|
11.333
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_+_20964414
|
11.281
|
|
|
|
|
chr12_-_123466195
|
11.256
|
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr1_+_44115828
|
11.103
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr5_+_180467224
|
10.978
|
NM_152547
|
BTNL9
|
butyrophilin-like 9
|
|
chr5_-_42811959
|
10.809
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr19_+_39899749
|
10.650
|
|
|
|
|
chr10_+_121578768
|
10.368
|
NM_001243194
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr1_-_160492982
|
10.272
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chrX_+_128913897
|
10.125
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr15_-_45670955
|
10.009
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chrX_+_128913881
|
9.996
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr2_+_113816214
|
9.908
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr7_+_23286277
|
9.861
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_-_89292396
|
9.808
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr7_+_105603656
|
9.731
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr7_+_87563432
|
9.678
|
NM_004194
NM_016351
NM_021721
NM_021722
NM_021723
|
ADAM22
|
ADAM metallopeptidase domain 22
|
|
chr7_+_23286401
|
9.432
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr17_-_26903854
|
9.228
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr16_+_30386097
|
9.219
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr1_+_22979681
|
9.177
|
NM_000491
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr1_+_158223926
|
9.161
|
NM_001763
|
CD1A
|
CD1a molecule
|
|
chr16_+_8814567
|
8.877
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr15_-_34212523
|
8.827
|
|
|
|
|
chr11_+_60048013
|
8.666
|
NM_001243266
NM_024021
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chr2_-_85895863
|
8.533
|
NM_000542
NM_198843
|
SFTPB
|
surfactant protein B
|
|
chr17_+_37824506
|
8.501
|
NM_002686
|
PNMT
|
phenylethanolamine N-methyltransferase
|
|
chr6_-_108145507
|
8.150
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr2_+_31456879
|
7.987
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr14_+_24616707
|
7.962
|
|
RNF31
|
ring finger protein 31
|
|
chr6_+_134210247
|
7.871
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr17_-_70588942
|
7.779
|
|
LOC100499467
|
uncharacterized LOC100499467
|
|
chr1_-_161279724
|
7.669
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr10_-_5541401
|
7.624
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr14_+_24616658
|
7.581
|
NM_017999
|
RNF31
|
ring finger protein 31
|
|
chr3_+_133495925
|
7.580
|
|
TF
|
transferrin
|
|
chr12_+_7055736
|
7.578
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr6_-_52628272
|
7.490
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr16_-_4852571
|
7.485
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr17_-_46115135
|
7.347
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr18_-_24442534
|
7.287
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr11_-_36619776
|
7.279
|
NM_000536
NM_001243785
NM_001243786
|
RAG2
|
recombination activating gene 2
|
|
chr6_+_134210264
|
7.246
|
|
TCF21
|
transcription factor 21
|
|
chr3_+_45067750
|
7.190
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr12_+_51318515
|
7.148
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr4_+_72204769
|
7.117
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr3_-_49314281
|
7.032
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr8_+_103563847
|
6.999
|
NM_024410
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr16_-_21314371
|
6.851
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr12_+_51318743
|
6.849
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr16_-_20702577
|
6.808
|
NM_052956
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1
|
|
chr5_-_149669187
|
6.790
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr1_-_147232684
|
6.780
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr1_+_209859524
|
6.691
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr9_-_90749899
|
6.643
|
NM_001166137
|
FAM75C2
|
family with sequence similarity 75, member C2
|
|
chr8_-_30891230
|
6.605
|
NM_001015508
|
PURG
|
purine-rich element binding protein G
|
|
chr1_+_162602308
|
6.586
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_-_153363544
|
6.509
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr8_-_120651019
|
6.269
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr8_-_110657745
|
6.143
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_33004771
|
6.132
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr14_+_70918873
|
6.080
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chrX_+_46940235
|
6.066
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr14_-_107131217
|
5.987
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr7_+_94539209
|
5.952
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr17_-_26903926
|
5.936
|
NM_005165
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr16_-_5115939
|
5.913
|
|
C16orf89
|
chromosome 16 open reading frame 89
|
|
chr3_-_187452669
|
5.861
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_+_7055753
|
5.763
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr7_+_123565908
|
5.752
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr6_+_150690027
|
5.716
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr1_+_13495253
|
5.639
|
NM_001045480
|
PRAMEF17
PRAMEF16
|
PRAME family member 17
PRAME family member 16
|
|
chr15_+_75105152
|
5.635
|
NM_021819
|
LMAN1L
|
lectin, mannose-binding, 1 like
|
|
chr1_-_182922504
|
5.618
|
NM_030933
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like
|
|
chr8_+_1952875
|
5.541
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr14_-_106774311
|
5.540
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr21_+_41239346
|
5.469
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chr6_-_52668599
|
5.445
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr7_+_123565285
|
5.412
|
NM_001174044
NM_003117
NM_153189
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr6_+_26156552
|
5.355
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr1_+_35225341
|
5.177
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr1_-_59147727
|
5.135
|
|
|
|
|
chrX_+_18709044
|
5.078
|
NM_006240
NM_152224
NM_152226
|
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1
|
|
chr6_-_99873129
|
5.062
|
NM_015491
NM_032870
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr9_-_77503009
|
5.060
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr10_-_123353330
|
4.945
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr22_+_45704821
|
4.893
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr9_+_102677457
|
4.876
|
|
STX17
|
syntaxin 17
|
|
chr15_+_42694587
|
4.870
|
NM_173088
|
CAPN3
|
calpain 3, (p94)
|
|
chr4_+_86699846
|
4.792
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr8_-_146202727
|
4.755
|
|
ZNF252
|
zinc finger protein 252
|
|
chr8_-_53322200
|
4.623
|
NM_014682
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein)
|
|
chr1_+_173793817
|
4.597
|
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial
|
|
chr14_-_106746056
|
4.566
|
|
|
|
|
chr2_+_113342171
|
4.530
|
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr11_+_118175294
|
4.511
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr22_+_22676814
|
4.472
|
|
|
|
|
chr2_-_43453724
|
4.462
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr15_-_43029318
|
4.431
|
NM_138477
|
CDAN1
|
congenital dyserythropoietic anemia, type I
|
|
chr10_-_47173917
|
4.374
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr22_+_23040394
|
4.332
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr4_+_113970784
|
4.302
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr1_+_43766565
|
4.288
|
NM_001253357
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr8_-_127570465
|
4.281
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr9_+_90532876
|
4.279
|
NM_001145124
|
FAM75C1
|
family with sequence similarity 75, member C1
|
|
chr9_-_73029539
|
4.267
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr2_-_202563353
|
4.266
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr17_-_43487747
|
4.235
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr5_+_35856990
|
4.186
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr17_+_7387773
|
4.124
|
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa
|
|
chr16_-_31100129
|
4.123
|
NM_001039503
|
PRSS53
|
protease, serine, 53
|
|
chr16_+_3451154
|
4.119
|
NM_001032292
NM_003450
|
ZNF174
|
zinc finger protein 174
|
|
chr1_-_11188800
|
4.118
|
|
|
|
|
chr16_+_90089007
|
4.116
|
NM_001481
|
GAS8
|
growth arrest-specific 8
|
|
chr11_-_118123001
|
4.105
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr13_+_96204941
|
4.094
|
NM_006984
|
CLDN10
|
claudin 10
|
|
chr12_-_54121203
|
4.052
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr6_-_49844808
|
4.039
|
NM_001205220
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr22_+_37678565
|
4.026
|
|
CYTH4
|
cytohesin 4
|
|
chr1_-_151319674
|
4.000
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr22_+_37678519
|
3.986
|
|
CYTH4
|
cytohesin 4
|
|
chr6_-_32908583
|
3.977
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr16_+_31366519
|
3.959
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr1_+_111770280
|
3.905
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr2_-_68384575
|
3.895
|
NM_138458
|
WDR92
|
WD repeat domain 92
|
|
chr10_-_123353771
|
3.852
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr5_-_147162251
|
3.847
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chrX_-_135749436
|
3.809
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_16758312
|
3.796
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_139943829
|
3.782
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr6_+_26156558
|
3.745
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr7_+_99621083
|
3.744
|
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr2_-_214016332
|
3.691
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr19_+_55014012
|
3.685
|
NM_002288
NM_021270
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2
|
|
chr12_+_69742130
|
3.673
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr1_-_111743261
|
3.652
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr14_+_23067128
|
3.593
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr5_-_149324282
|
3.564
|
NM_000440
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha
|
|
chr9_-_21202201
|
3.535
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr3_-_146213721
|
3.531
|
NM_001199979
|
PLSCR2
|
phospholipid scramblase 2
|
|
chrX_+_65382627
|
3.511
|
|
HEPH
|
hephaestin
|
|
chr5_+_137203559
|
3.496
|
|
MYOT
|
myotilin
|
|
chr19_+_58570606
|
3.452
|
NM_001164527
NM_001164529
NM_001164530
NM_003436
NM_007134
|
ZNF135
|
zinc finger protein 135
|
|
chr19_+_54926721
|
3.412
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr6_-_134496833
|
3.371
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_-_62009620
|
3.356
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr1_+_159173802
|
3.349
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr17_-_29641094
|
3.317
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_+_89829435
|
3.276
|
NM_198460
|
GBP6
|
guanylate binding protein family, member 6
|
|
chr7_-_124405680
|
3.244
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr10_+_102672685
|
3.233
|
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr17_+_77751929
|
3.218
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr16_-_20338808
|
3.195
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr4_-_76823575
|
3.193
|
NM_006239
|
PPEF2
|
protein phosphatase, EF-hand calcium binding domain 2
|
|
chr9_+_131388167
|
3.179
|
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr3_-_194090459
|
3.174
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chr11_-_59633942
|
3.169
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr1_-_111743297
|
3.169
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr17_+_11786218
|
3.123
|
NM_004662
|
DNAH9
|
dynein, axonemal, heavy chain 9
|
|
chr12_+_50898744
|
3.086
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr4_-_95263986
|
3.078
|
NM_014485
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr11_-_118047145
|
3.062
|
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chrX_+_48114796
|
3.049
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr3_+_155838336
|
3.030
|
NM_172160
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr17_+_21308447
|
2.961
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chrX_-_100548044
|
2.960
|
NM_024885
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
|
|
chr14_-_45722604
|
2.943
|
NM_018353
|
MIS18BP1
|
MIS18 binding protein 1
|
|
chr12_-_16757944
|
2.922
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_+_50472856
|
2.907
|
|
SIGLEC16
|
sialic acid binding Ig-like lectin 16 (gene/pseudogene)
|
|
chr6_-_89927495
|
2.906
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr1_+_162602198
|
2.865
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr17_+_7960201
|
2.840
|
|
|
|
|
chr3_+_35722428
|
2.822
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr17_-_8815799
|
2.815
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|