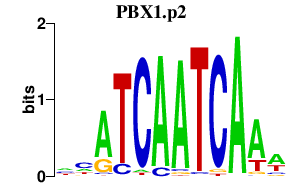
|
chr1_-_28241233
|
39.561
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr5_+_162864571
|
39.419
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr9_+_140135698
|
34.638
|
NM_006088
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chrX_+_119737743
|
34.563
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr9_+_140135738
|
32.071
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr17_+_8191814
|
32.011
|
NM_001177801
NM_001177802
NM_016492
|
RANGRF
|
RAN guanine nucleotide release factor
|
|
chr7_-_7680048
|
28.893
|
|
RPA3
|
replication protein A3, 14kDa
|
|
chr4_-_174255454
|
27.727
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chrX_+_119737905
|
27.551
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr9_+_140135742
|
26.285
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chrX_+_119737984
|
25.573
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr12_+_64803741
|
25.371
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr3_-_32544358
|
24.483
|
NM_017801
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr12_+_49658862
|
23.896
|
NM_032704
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr3_+_172468469
|
23.418
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr6_+_111195986
|
23.367
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr6_-_75965845
|
22.639
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr1_-_28240953
|
22.607
|
|
RPA2
|
replication protein A2, 32kDa
|
|
chr12_+_49658865
|
22.297
|
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr2_+_216986817
|
21.728
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chrX_+_123234462
|
21.067
|
|
STAG2
|
stromal antigen 2
|
|
chr5_-_89705477
|
20.806
|
NM_004365
|
CETN3
|
centrin, EF-hand protein, 3
|
|
chr15_-_64673569
|
20.598
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr10_+_102123420
|
20.238
|
|
|
|
|
chr6_-_27114577
|
19.844
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr17_-_38821286
|
19.509
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr6_+_149721494
|
19.205
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr20_+_44441334
|
18.793
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr6_+_27114860
|
18.726
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr14_+_64971504
|
18.509
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr5_+_44808967
|
18.489
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr10_+_14880335
|
17.080
|
|
HSPA14
|
heat shock 70kDa protein 14
|
|
chr3_-_129158675
|
17.074
|
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chr3_-_32544319
|
16.944
|
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr3_-_129158849
|
16.754
|
NM_003925
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chr3_-_129158779
|
16.686
|
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chr3_-_33481877
|
16.463
|
NM_001128161
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr1_+_155278727
|
16.421
|
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr3_-_33481862
|
16.149
|
NM_001128160
NM_014517
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr19_+_14640413
|
15.973
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr1_+_155278677
|
15.937
|
NM_001135821
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr10_+_14880343
|
15.874
|
|
HSPA14
|
heat shock 70kDa protein 14
|
|
chr3_+_38206968
|
15.805
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr1_-_115268890
|
15.203
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr19_+_54619149
|
15.115
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr3_+_156394452
|
14.967
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr3_+_179280738
|
14.858
|
|
ACTL6A
|
actin-like 6A
|
|
chr8_-_101965554
|
14.837
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr5_-_179044835
|
14.764
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr13_+_53035227
|
14.732
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr10_+_14880137
|
14.528
|
NM_016299
|
HSPA14
|
heat shock 70kDa protein 14
|
|
chr5_+_145826866
|
14.415
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr8_-_101965096
|
14.375
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr12_-_120638894
|
14.247
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr7_-_140179224
|
14.197
|
|
MKRN1
|
makorin ring finger protein 1
|
|
chr20_+_44441247
|
14.104
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr19_+_14640406
|
13.656
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr2_-_47134945
|
13.253
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr12_-_120638909
|
13.023
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr8_-_25315941
|
12.622
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr1_+_151227209
|
12.507
|
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr11_+_95523608
|
12.426
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr19_+_41770273
|
12.285
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_-_200322699
|
12.135
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr19_+_14640403
|
12.055
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr20_-_45976626
|
11.874
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr3_+_179280746
|
11.816
|
|
ACTL6A
|
actin-like 6A
|
|
chr14_+_23938883
|
11.722
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr6_+_26104103
|
11.716
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr8_+_107738239
|
11.493
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr3_-_148804122
|
11.413
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr14_-_54908035
|
11.305
|
NM_005776
|
CNIH
|
cornichon homolog (Drosophila)
|
|
chr7_-_148725764
|
11.294
|
NM_004911
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr5_+_43602738
|
11.278
|
NM_012343
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr20_+_30326903
|
11.249
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr19_+_14640363
|
10.494
|
NM_138501
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr5_+_151151514
|
10.464
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr1_-_149399159
|
10.288
|
NM_001024599
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr1_+_151227188
|
10.282
|
NM_002810
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr3_+_179280686
|
10.282
|
NM_004301
NM_177989
NM_178042
|
ACTL6A
|
actin-like 6A
|
|
chr1_-_3713048
|
10.212
|
NM_020710
|
LRRC47
|
leucine rich repeat containing 47
|
|
chr12_-_121790165
|
10.201
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr5_+_43603216
|
9.825
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr7_-_140179133
|
9.776
|
|
MKRN1
|
makorin ring finger protein 1
|
|
chr22_-_43411129
|
9.721
|
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr22_-_43411156
|
9.652
|
NM_001184970
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr7_-_148725731
|
9.363
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr1_+_35851101
|
9.338
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr12_+_28410132
|
9.328
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr11_+_67798077
|
9.251
|
NM_002496
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
|
|
chr5_+_43603312
|
9.209
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr8_+_98788014
|
9.165
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr14_-_55658205
|
8.970
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr3_+_136581007
|
8.832
|
NM_006153
|
NCK1
|
NCK adaptor protein 1
|
|
chr1_+_145663156
|
8.625
|
|
RNF115
|
ring finger protein 115
|
|
chr1_+_155278538
|
8.569
|
NM_001135822
NM_001242824
NM_001242825
NM_002004
|
FDPS
|
farnesyl diphosphate synthase
|
|
chrX_-_10645726
|
8.527
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr19_+_54619114
|
8.351
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr2_-_219134846
|
8.332
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr1_+_149858466
|
8.319
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr1_+_32645322
|
8.311
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr2_+_171784947
|
8.248
|
NM_001201428
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr1_-_241683060
|
8.012
|
NM_000143
|
FH
|
fumarate hydratase
|
|
chrX_-_23761396
|
7.960
|
NM_001033583
NM_001037171
|
ACOT9
|
acyl-CoA thioesterase 9
|
|
chr12_-_121790141
|
7.936
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr19_+_41770285
|
7.872
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr7_-_7758237
|
7.867
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr19_-_39881631
|
7.740
|
NM_019088
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr3_+_171757432
|
7.727
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr12_-_56679729
|
7.667
|
|
CS
|
citrate synthase
|
|
chr1_-_42801529
|
7.662
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chrX_+_70503496
|
7.653
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr1_-_241683011
|
7.632
|
|
FH
|
fumarate hydratase
|
|
chr5_+_151151462
|
7.575
|
NM_005754
NM_198395
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr12_+_21168629
|
7.529
|
NM_001009562
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional)
|
|
chr16_-_58663734
|
7.499
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr3_+_136581074
|
7.474
|
|
NCK1
|
NCK adaptor protein 1
|
|
chr1_-_156308047
|
7.468
|
|
CCT3
|
chaperonin containing TCP1, subunit 3 (gamma)
|
|
chr2_+_12856813
|
7.443
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr12_-_121790240
|
7.438
|
NM_016237
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr19_+_2269515
|
7.429
|
NM_004152
|
OAZ1
SPPL2B
|
ornithine decarboxylase antizyme 1
signal peptide peptidase-like 2B
|
|
chr9_+_91926109
|
7.423
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr11_+_95523809
|
7.417
|
|
CEP57
|
centrosomal protein 57kDa
|
|
chr19_+_49375679
|
7.416
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr1_+_236557643
|
7.385
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr16_+_69220940
|
7.337
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr4_-_48782281
|
7.328
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr5_+_179132676
|
7.289
|
|
CANX
|
calnexin
|
|
chr1_+_214776494
|
7.275
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr3_+_136649316
|
7.262
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr19_-_14247400
|
7.244
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr19_+_49375677
|
7.230
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr6_-_42713822
|
7.201
|
|
TBCC
|
tubulin folding cofactor C
|
|
chr12_-_120638912
|
7.201
|
NM_001002
NM_053275
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr3_-_160283166
|
7.184
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr19_-_14640048
|
7.181
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr2_-_219134865
|
7.155
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr17_-_29151724
|
7.150
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr17_-_29151638
|
7.143
|
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr19_+_2269526
|
7.106
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr9_+_79968344
|
7.029
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr12_-_14956408
|
7.006
|
|
WBP11
|
WW domain binding protein 11
|
|
chr5_+_151151516
|
6.976
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr4_+_83956238
|
6.956
|
NM_016129
|
COPS4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis)
|
|
chr2_-_219134795
|
6.905
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr11_-_57195052
|
6.904
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr11_+_69467210
|
6.853
|
|
CCND1
|
cyclin D1
|
|
chr19_+_49375638
|
6.845
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr2_-_61765394
|
6.826
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chrX_+_70503469
|
6.725
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr6_+_160183510
|
6.682
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chrX_-_107334749
|
6.666
|
NM_002814
NM_170750
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10
|
|
chr2_+_191745547
|
6.657
|
|
GLS
|
glutaminase
|
|
chr14_-_75530716
|
6.655
|
NM_001107
NM_203488
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type
|
|
chr17_+_73663398
|
6.650
|
NM_013260
|
SAP30BP
|
SAP30 binding protein
|
|
chr4_-_72649734
|
6.583
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chrX_+_47004657
|
6.553
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr19_-_18433847
|
6.539
|
|
LSM4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chrX_+_70503512
|
6.404
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr1_-_156307971
|
6.395
|
|
CCT3
|
chaperonin containing TCP1, subunit 3 (gamma)
|
|
chr19_+_41770057
|
6.317
|
NM_007040
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chrX_+_47004663
|
6.295
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr9_+_131447364
|
6.260
|
NM_001248000
|
SET
|
SET nuclear oncogene
|
|
chr2_-_174829951
|
6.232
|
|
SP3
|
Sp3 transcription factor
|
|
chrX_+_47004676
|
6.210
|
|
RBM10
|
RNA binding motif protein 10
|
|
chrX_+_47004693
|
6.200
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr6_-_42713837
|
6.173
|
NM_003192
|
TBCC
|
tubulin folding cofactor C
|
|
chr18_+_77866914
|
6.173
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr19_+_2269529
|
6.117
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr17_-_46622274
|
6.032
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chrX_+_47004665
|
6.029
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr15_+_59397276
|
6.027
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chrX_+_47004633
|
5.940
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr20_+_44441671
|
5.912
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr12_-_14956367
|
5.904
|
NM_016312
|
WBP11
|
WW domain binding protein 11
|
|
chr12_-_120315074
|
5.896
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr3_-_160283071
|
5.880
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chrX_+_47004609
|
5.818
|
NM_001204466
NM_001204467
NM_001204468
NM_005676
NM_152856
|
RBM10
|
RNA binding motif protein 10
|
|
chr3_-_121553907
|
5.784
|
NM_001023570
NM_001023571
|
IQCB1
|
IQ motif containing B1
|
|
chrX_+_47004670
|
5.782
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr4_-_84035888
|
5.768
|
|
PLAC8
|
placenta-specific 8
|
|
chr1_+_110881944
|
5.696
|
NM_001201545
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr9_+_36190885
|
5.689
|
|
CLTA
|
clathrin, light chain A
|
|
chr2_-_61765367
|
5.675
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr19_-_39881503
|
5.667
|
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr17_-_1588098
|
5.635
|
|
PRPF8
|
PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae)
|
|
chr17_-_79818318
|
5.618
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr22_-_30642683
|
5.611
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr2_-_61765345
|
5.610
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr3_-_121553836
|
5.596
|
|
IQCB1
|
IQ motif containing B1
|
|
chr22_-_17640153
|
5.592
|
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr19_+_54704659
|
5.506
|
NM_001013
|
RPS9
|
ribosomal protein S9
|
|
chr12_-_54653317
|
5.501
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr3_-_160283311
|
5.475
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr18_-_47018842
|
5.424
|
NM_001199356
NM_000985
NM_001035006
NM_001199340
NM_001199345
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr10_+_70660955
|
5.382
|
NM_024045
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50
|
|
chr14_+_93673583
|
5.353
|
|
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative)
|
|
chr14_+_96001297
|
5.326
|
NM_016417
|
GLRX5
|
glutaredoxin 5
|
|
chr20_+_18488525
|
5.306
|
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chrX_+_70503534
|
5.260
|
|
NONO
|
non-POU domain containing, octamer-binding
|