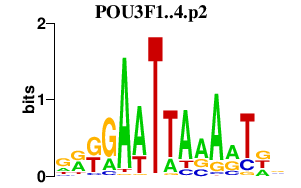
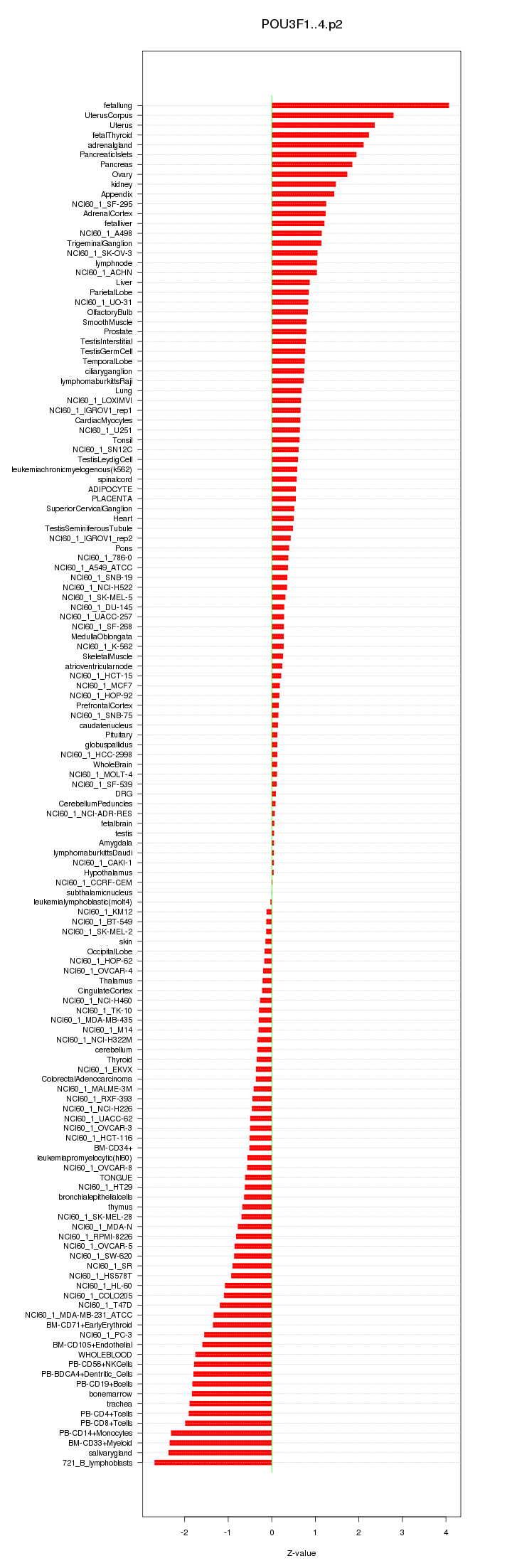
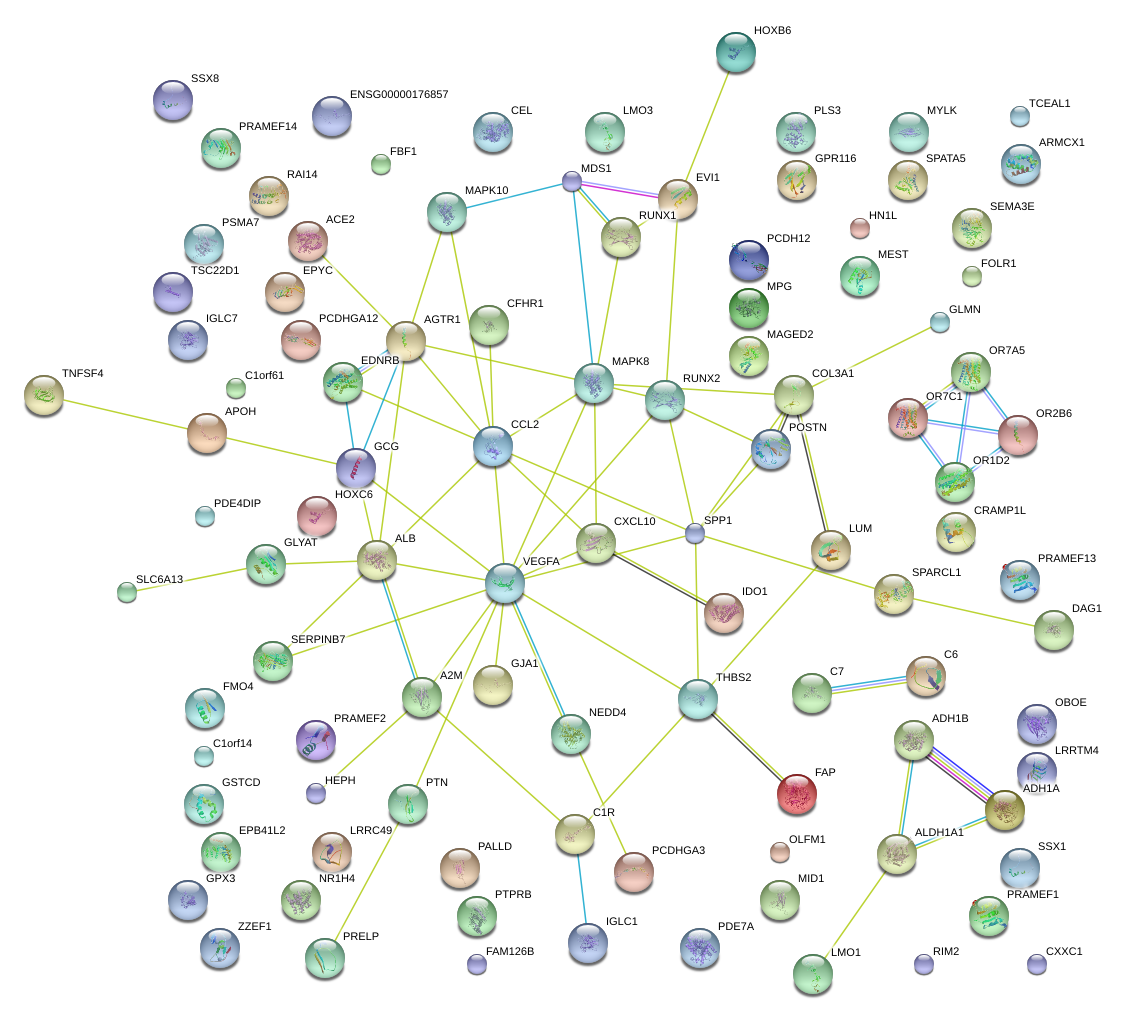
|
chr5_+_150399980
|
19.537
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr5_+_40909617
|
17.330
|
|
C7
|
complement component 7
|
|
chr5_+_40909558
|
16.172
|
NM_000587
|
C7
|
complement component 7
|
|
chr2_+_189839113
|
15.427
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839132
|
13.964
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_+_121756744
|
12.725
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr12_-_91505215
|
11.909
|
|
LUM
|
lumican
|
|
chr4_+_74270832
|
11.857
|
|
ALB
|
albumin
|
|
chr4_+_88896868
|
11.697
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_189839078
|
10.990
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_-_46889620
|
10.948
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr4_+_88896838
|
10.228
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_123339178
|
10.149
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_16759939
|
9.340
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16759733
|
8.949
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_65384078
|
8.165
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr4_-_88450601
|
7.651
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr4_+_74269971
|
7.288
|
NM_000477
|
ALB
|
albumin
|
|
chr12_-_9268513
|
6.867
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr8_+_39771327
|
6.445
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr12_-_9268475
|
6.342
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr4_+_88896801
|
6.191
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chrX_+_100805415
|
6.092
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr4_+_169418167
|
5.986
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr5_+_140729723
|
5.898
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr12_-_7245048
|
5.850
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr15_-_56209305
|
5.607
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr17_+_32582295
|
5.508
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_-_131291480
|
5.443
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr5_-_41213513
|
5.429
|
NM_000065
|
C6
|
complement component 6
|
|
chr4_-_100242487
|
5.370
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr1_+_171283372
|
5.356
|
NM_002022
|
FMO4
|
flavin containing monooxygenase 4
|
|
chr2_-_163008913
|
5.275
|
NM_002054
|
GCG
|
glucagon
|
|
chrX_+_102883891
|
5.250
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr3_-_123339423
|
5.237
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_168865521
|
5.226
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_+_148447966
|
5.004
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr17_-_64225496
|
4.944
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr17_-_46682333
|
4.659
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr18_+_61442611
|
4.625
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr16_+_1728290
|
4.608
|
|
HN1L
|
hematological and neurological expressed 1-like
|
|
chr17_-_2996289
|
4.590
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr16_+_4845265
|
4.516
|
NM_001253793
NM_001253794
|
LOC440335
|
uncharacterized LOC440335
|
|
chr11_+_71903172
|
4.449
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr7_-_137028433
|
4.405
|
|
PTN
|
pleiotrophin
|
|
chr12_-_71031174
|
4.358
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr7_-_137028370
|
4.295
|
|
PTN
|
pleiotrophin
|
|
chr6_-_169654047
|
4.285
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr2_-_163099877
|
4.035
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr6_+_43738394
|
3.956
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr7_-_83278478
|
3.824
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr9_+_137967366
|
3.790
|
|
OLFM1
|
olfactomedin 1
|
|
chr13_-_45011389
|
3.787
|
NM_001243797
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr5_+_34684611
|
3.745
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr6_+_27925018
|
3.684
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr4_-_87028805
|
3.609
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr4_+_123844210
|
3.558
|
NM_145207
|
SPATA5
|
spermatogenesis associated 5
|
|
chr9_-_75568232
|
3.548
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr15_+_71184781
|
3.351
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr1_-_156399183
|
3.333
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr4_+_106631966
|
3.325
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr1_-_173174611
|
3.322
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr12_+_21168629
|
3.279
|
NM_001009562
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional)
|
|
chr12_+_100897137
|
3.250
|
NM_001206992
NM_001206993
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr5_-_141338626
|
3.203
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr12_-_91398744
|
3.164
|
|
EPYC
|
epiphycan
|
|
chr1_-_144997110
|
3.150
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr5_+_140810181
|
3.136
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr13_-_78492906
|
3.114
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr8_+_104831415
|
3.113
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr11_-_58499368
|
3.111
|
NM_005838
NM_201648
|
GLYAT
|
glycine-N-acyltransferase
|
|
chr9_+_137967267
|
3.103
|
|
OLFM1
|
olfactomedin 1
|
|
chr12_-_9268522
|
3.072
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_196788860
|
3.070
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr10_+_102123420
|
3.064
|
|
|
|
|
chrX_-_15620191
|
2.960
|
NM_021804
|
ACE2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr7_+_45933096
|
2.952
|
|
|
|
|
chrX_+_54834775
|
2.887
|
NM_014599
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr1_-_182922504
|
2.856
|
NM_030933
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like
|
|
chr13_-_38172862
|
2.809
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr8_-_66701175
|
2.807
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr12_-_371994
|
2.800
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chrX_+_48114796
|
2.770
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chrX_+_114827818
|
2.696
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr12_+_54422193
|
2.644
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr12_-_16759430
|
2.634
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr21_-_36421461
|
2.630
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_203444955
|
2.601
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr5_+_140792507
|
2.597
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr5_+_140810126
|
2.596
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr19_-_14939275
|
2.553
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr1_-_13452655
|
2.534
|
NM_001024661
NM_001099854
|
PRAMEF13
PRAMEF14
|
PRAME family member 13
PRAME family member 14
|
|
chr2_-_77749335
|
2.523
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr2_-_201936386
|
2.508
|
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr7_-_83270746
|
2.501
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr7_+_130126035
|
2.495
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr3_+_49568370
|
2.491
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chrX_-_10535476
|
2.456
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_+_64708614
|
2.442
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr11_-_85469043
|
2.441
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_147211141
|
2.397
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr6_+_46761093
|
2.393
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr8_-_17579729
|
2.378
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr4_-_88450243
|
2.362
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr18_+_61575203
|
2.280
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chrX_+_91089658
|
2.271
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr3_+_11267666
|
2.256
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr17_-_66951381
|
2.241
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_-_114343052
|
2.228
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr10_-_5060191
|
2.202
|
NM_001354
NM_205845
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr7_-_126883568
|
2.198
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr7_-_80141241
|
2.191
|
NM_001102386
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3
|
|
chr1_-_163113938
|
2.176
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr10_+_95848811
|
2.131
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr5_+_140743755
|
2.114
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr15_-_23932410
|
2.106
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr10_-_69425356
|
2.093
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr9_-_21187529
|
2.087
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr11_-_71781091
|
2.055
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr13_-_36429798
|
2.042
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chrX_+_67913479
|
2.032
|
NM_001142504
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr1_+_15986363
|
2.027
|
NM_006511
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1
|
|
chr5_-_58652671
|
2.026
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_124405680
|
2.013
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr7_-_137028275
|
2.002
|
|
PTN
|
pleiotrophin
|
|
chr13_+_24844846
|
1.964
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr13_-_78493902
|
1.961
|
NM_001201397
|
EDNRB
|
endothelin receptor type B
|
|
chr12_+_57984992
|
1.936
|
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr4_-_72649734
|
1.910
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr6_+_28234787
|
1.885
|
NM_001023560
NM_001111039
NM_152736
|
ZNF187
|
zinc finger protein 187
|
|
chr16_-_90039239
|
1.875
|
NM_145039
|
CENPBD1
|
CENPB DNA-binding domains containing 1
|
|
chr4_+_88720701
|
1.837
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr13_+_102104941
|
1.831
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr19_+_56186569
|
1.827
|
|
EPN1
|
epsin 1
|
|
chr3_-_99594915
|
1.824
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr5_+_149340287
|
1.808
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr16_+_6823809
|
1.802
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr11_-_8954534
|
1.792
|
NM_020643
|
C11orf16
|
chromosome 11 open reading frame 16
|
|
chr1_-_93399531
|
1.786
|
|
|
|
|
chr20_+_9458460
|
1.759
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr17_+_42925278
|
1.759
|
NM_016438
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B
|
|
chr6_+_27107060
|
1.739
|
NM_003495
|
HIST2H4B
HIST1H4I
|
histone cluster 2, H4b
histone cluster 1, H4i
|
|
chr4_-_155533800
|
1.731
|
|
FGG
|
fibrinogen gamma chain
|
|
chr3_-_194991778
|
1.722
|
NM_152531
|
XXYLT1
|
xyloside xylosyltransferase 1
|
|
chr5_+_71502700
|
1.711
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_157002705
|
1.699
|
|
|
|
|
chr18_-_53068755
|
1.695
|
|
TCF4
|
transcription factor 4
|
|
chr3_+_121613170
|
1.691
|
NM_001145998
NM_021082
|
SLC15A2
|
solute carrier family 15 (H+/peptide transporter), member 2
|
|
chr4_-_110723127
|
1.690
|
|
CFI
|
complement factor I
|
|
chr1_+_150230168
|
1.687
|
NM_012113
|
CA14
|
carbonic anhydrase XIV
|
|
chrX_-_137821514
|
1.684
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr3_-_46718898
|
1.679
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr5_-_75919051
|
1.672
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr9_-_111618209
|
1.670
|
NM_006686
|
ACTL7B
|
actin-like 7B
|
|
chr15_+_42066690
|
1.639
|
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr3_+_153838791
|
1.633
|
NM_001251962
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr7_-_137028533
|
1.630
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr12_+_12224401
|
1.628
|
NM_030766
NM_138722
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator)
|
|
chr15_+_59908908
|
1.610
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr3_-_114102488
|
1.596
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_+_69307033
|
1.592
|
NM_001184779
NM_024505
|
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr5_+_140771482
|
1.589
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr4_+_71108304
|
1.589
|
NM_005212
|
CSN3
|
casein kappa
|
|
chrX_+_110366304
|
1.587
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr11_-_118123001
|
1.556
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr22_+_18043147
|
1.550
|
NM_031481
|
SLC25A18
|
solute carrier family 25 (mitochondrial carrier), member 18
|
|
chr1_+_196912910
|
1.535
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr1_+_182419255
|
1.532
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr1_+_12916940
|
1.526
|
NM_023014
|
PRAMEF2
|
PRAME family member 2
|
|
chr4_-_100273831
|
1.505
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr6_-_49834188
|
1.484
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr21_-_39956823
|
1.484
|
NM_001243429
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr19_-_4400357
|
1.480
|
NM_001199944
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr7_+_102023016
|
1.475
|
|
|
|
|
chr10_+_102121371
|
1.465
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr1_+_78470593
|
1.458
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr10_+_15085894
|
1.457
|
NM_001039702
NM_018324
|
OLAH
|
oleoyl-ACP hydrolase
|
|
chr21_-_36421195
|
1.456
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_138820580
|
1.454
|
|
|
|
|
chr19_+_16830786
|
1.432
|
NM_001007525
|
NWD1
|
NACHT and WD repeat domain containing 1
|
|
chr4_-_110723334
|
1.427
|
NM_000204
|
CFI
|
complement factor I
|
|
chr7_-_87342569
|
1.419
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr2_-_31352489
|
1.416
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr2_-_209010876
|
1.415
|
NM_005210
|
CRYGB
|
crystallin, gamma B
|
|
chr8_-_13372183
|
1.412
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_146202727
|
1.409
|
|
ZNF252
|
zinc finger protein 252
|
|
chr12_-_58135853
|
1.386
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chrX_+_105937067
|
1.385
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr17_+_28268622
|
1.380
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr9_+_131388167
|
1.369
|
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr6_-_136788012
|
1.367
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr16_+_81272293
|
1.364
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chrX_-_32430370
|
1.363
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr2_+_152214105
|
1.358
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chrX_+_144908927
|
1.355
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr4_-_26491888
|
1.353
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr9_+_117904096
|
1.347
|
NM_017418
|
DEC1
|
deleted in esophageal cancer 1
|
|
chr1_+_12851545
|
1.329
|
NM_023013
|
PRAMEF1
|
PRAME family member 1
|
|
chr5_+_81267797
|
1.328
|
NM_001131028
NM_031482
|
ATG10
|
ATG10 autophagy related 10 homolog (S. cerevisiae)
|
|
chr4_+_156824846
|
1.322
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|