|
chr15_+_74422920
|
26.742
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr13_-_45010974
|
25.609
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr15_+_74422742
|
24.837
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr10_+_5005685
|
24.518
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr13_-_45010994
|
23.675
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_45010937
|
23.424
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr22_+_22735134
|
19.958
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr10_+_5005601
|
15.902
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_5005453
|
15.600
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr22_+_22786283
|
15.530
|
|
|
|
|
chrX_+_135278912
|
15.232
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
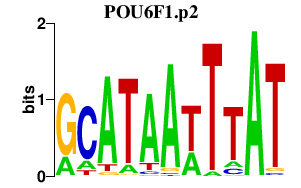
chr12_-_51611476
|
13.727
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr13_-_45011389
|
13.561
|
NM_001243797
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr12_-_91572328
|
11.820
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr9_-_95166827
|
11.589
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr2_+_113816214
|
10.796
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr13_-_45011059
|
10.486
|
NM_006022
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr4_+_70861647
|
10.476
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chrX_+_67913479
|
10.441
|
NM_001142504
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr22_+_39101954
|
10.186
|
|
GTPBP1
|
GTP binding protein 1
|
|
chr3_-_13057130
|
9.934
|
|
|
|
|
chr22_+_22735202
|
9.852
|
|
|
|
|
chr7_-_100026284
|
9.632
|
NM_017984
|
ZCWPW1
|
zinc finger, CW type with PWWP domain 1
|
|
chr1_-_116311161
|
9.587
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr7_-_64023444
|
9.583
|
|
|
|
|
chr1_-_116311330
|
9.541
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr19_+_39899749
|
9.507
|
|
|
|
|
chrX_+_65382432
|
9.451
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr9_+_34458810
|
9.147
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr1_-_116311398
|
8.991
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr7_+_39017608
|
8.844
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr22_+_22735207
|
8.775
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr10_-_5045967
|
8.374
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr7_-_27183225
|
8.057
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_-_137475076
|
8.048
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr3_-_57678594
|
7.580
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr18_+_47088400
|
6.725
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chrX_+_91090459
|
6.304
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr17_-_2996289
|
5.966
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr3_+_46919235
|
5.846
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr17_-_48945338
|
5.661
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr19_-_49658645
|
5.588
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr16_-_31439650
|
5.523
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr3_-_123339178
|
5.510
|
|
MYLK
|
myosin light chain kinase
|
|
chr1_+_104159953
|
5.496
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr17_+_17608163
|
5.482
|
|
|
|
|
chr6_+_27925018
|
5.459
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr6_+_72926144
|
5.351
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_-_83393436
|
5.317
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_-_89664510
|
5.311
|
NM_052941
|
GBP4
|
guanylate binding protein 4
|
|
chr13_-_45048436
|
5.245
|
NM_001243798
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr16_-_70834884
|
5.152
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr14_+_39703119
|
5.123
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr16_-_70835044
|
5.057
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr17_-_67240955
|
5.048
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr4_-_87028805
|
4.986
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr22_+_40322609
|
4.939
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr16_-_70835061
|
4.854
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr11_-_59633942
|
4.825
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr5_+_125759076
|
4.818
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr22_-_22863504
|
4.736
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr1_+_196621007
|
4.707
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr17_-_66951381
|
4.700
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr19_-_33360641
|
4.679
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr2_-_216241321
|
4.652
|
|
FN1
|
fibronectin 1
|
|
chr12_+_21284127
|
4.638
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr12_-_16760935
|
4.533
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr22_+_22758538
|
4.478
|
|
|
|
|
chr6_+_54173202
|
4.405
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr17_+_41857802
|
4.363
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr4_+_175839508
|
4.303
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr16_+_2653350
|
4.286
|
|
LOC652276
|
potassium channel tetramerisation domain containing 5 pseudogene
|
|
chr6_+_72926462
|
4.267
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_+_142829169
|
4.264
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr16_-_32688052
|
4.207
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr10_+_96443250
|
4.157
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr6_-_136847078
|
4.144
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr19_-_21950322
|
4.095
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr6_+_144612872
|
4.069
|
NM_007124
|
UTRN
|
utrophin
|
|
chr5_+_125758818
|
4.067
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr16_+_33261514
|
4.059
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr19_-_18632857
|
4.026
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr1_-_104239071
|
4.015
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr4_+_78432905
|
3.887
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr2_-_1926501
|
3.874
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_70835067
|
3.843
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr4_+_71587674
|
3.785
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr21_+_19289656
|
3.771
|
NM_001204175
NM_001204176
NM_001204177
NM_001204178
|
CHODL
|
chondrolectin
|
|
chr5_+_92918924
|
3.747
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr9_-_215892
|
3.730
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr2_-_169887832
|
3.717
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr4_-_122085475
|
3.668
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr16_+_33204979
|
3.631
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr16_+_81272293
|
3.614
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr1_+_153003677
|
3.583
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr8_+_19796581
|
3.465
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr5_-_160279045
|
3.459
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr10_+_97889467
|
3.393
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr12_-_28124915
|
3.377
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr2_+_233390869
|
3.352
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr2_+_89185055
|
3.316
|
|
|
|
|
chr19_+_24269877
|
3.307
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr6_-_49834188
|
3.296
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr16_-_70835060
|
3.270
|
NM_018052
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr19_-_18632886
|
3.266
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr7_+_100027253
|
3.260
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr14_+_70918873
|
3.241
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr7_-_27159213
|
3.240
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr5_-_75919051
|
3.168
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr14_-_107013474
|
3.106
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr18_+_61144143
|
3.086
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr11_+_71927818
|
3.080
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr11_+_5009420
|
3.023
|
NM_021801
|
MMP26
|
matrix metallopeptidase 26
|
|
chr9_-_95298251
|
3.016
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr1_-_104238785
|
2.972
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr13_-_46679143
|
2.940
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr3_+_186330859
|
2.888
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr9_+_19290748
|
2.865
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr10_+_104503724
|
2.861
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr17_-_10421858
|
2.845
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr17_-_39041436
|
2.811
|
NM_019010
|
KRT20
|
keratin 20
|
|
chr1_+_46379258
|
2.801
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr11_+_74699939
|
2.752
|
NM_006656
|
NEU3
|
sialidase 3 (membrane sialidase)
|
|
chr2_-_89619820
|
2.688
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr2_+_176969233
|
2.659
|
|
|
|
|
chr22_+_23040394
|
2.655
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr12_-_7848324
|
2.637
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chrX_+_65382627
|
2.636
|
|
HEPH
|
hephaestin
|
|
chr4_+_88720701
|
2.632
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr6_+_100054649
|
2.626
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr8_+_17396285
|
2.570
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr6_+_154407641
|
2.561
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr12_-_4488707
|
2.538
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr7_-_99766191
|
2.483
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr6_+_30130982
|
2.467
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr1_-_163113938
|
2.432
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_+_26503973
|
2.408
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr2_+_210444154
|
2.396
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_+_104198140
|
2.341
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr8_-_67090690
|
2.334
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr1_-_193155723
|
2.332
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr7_-_83278478
|
2.323
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr1_+_15272414
|
2.286
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr3_-_18480203
|
2.281
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr6_+_134758840
|
2.270
|
|
LOC154092
|
uncharacterized LOC154092
|
|
chr7_-_50628744
|
2.244
|
NM_000790
NM_001242886
NM_001242887
NM_001242888
NM_001242889
NM_001242890
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr4_+_62362838
|
2.232
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr4_+_41614911
|
2.229
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_+_41583407
|
2.219
|
NM_080817
|
GPR82
|
G protein-coupled receptor 82
|
|
chr12_+_51985016
|
2.216
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr5_-_138211054
|
2.203
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chrX_-_32173585
|
2.162
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr14_-_101036130
|
2.135
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr1_+_162332772
|
2.134
|
NM_001126060
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr17_-_56492938
|
2.115
|
|
RNF43
|
ring finger protein 43
|
|
chr10_-_15902455
|
2.098
|
NM_024948
|
FAM188A
|
family with sequence similarity 188, member A
|
|
chr6_-_49754899
|
2.096
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr5_-_147162251
|
2.015
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr22_+_22712091
|
1.982
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr17_+_20771745
|
1.956
|
|
|
|
|
chr5_+_147443534
|
1.944
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr1_+_226012974
|
1.914
|
NM_000120
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr1_+_13521962
|
1.908
|
NM_001100114
|
PRAMEF21
|
PRAME family member 21
|
|
chr5_+_140220811
|
1.898
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr7_-_99766238
|
1.880
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr19_-_22379752
|
1.819
|
NM_001001411
|
ZNF676
|
zinc finger protein 676
|
|
chr7_+_48211054
|
1.817
|
NM_152701
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13
|
|
chr15_+_52121855
|
1.795
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr15_+_93443418
|
1.788
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr19_+_15852200
|
1.769
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr1_+_18081807
|
1.760
|
NM_030812
|
ACTL8
|
actin-like 8
|
|
chr3_+_186330711
|
1.758
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr7_+_138818489
|
1.758
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr1_-_117746457
|
1.718
|
NM_001253849
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr1_+_104292439
|
1.710
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr1_-_13673510
|
1.688
|
NM_001099854
|
PRAMEF14
|
PRAME family member 14
|
|
chr14_-_107013122
|
1.665
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_+_93535819
|
1.660
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr2_-_183387063
|
1.643
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr7_-_100425035
|
1.639
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr1_+_160336856
|
1.635
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr11_+_124824012
|
1.630
|
NM_025004
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr11_-_8190535
|
1.607
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr3_+_142442855
|
1.597
|
|
|
|
|
chr14_-_106926393
|
1.595
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr13_-_52980628
|
1.553
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr13_+_52586516
|
1.534
|
NM_001004127
|
ALG11
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
|
|
chr6_-_112575708
|
1.526
|
|
LAMA4
|
laminin, alpha 4
|
|
chr4_+_95376396
|
1.525
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr5_-_134871638
|
1.495
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr1_+_196743929
|
1.482
|
NM_001166624
NM_021023
|
CFHR3
|
complement factor H-related 3
|
|
chr12_-_11062160
|
1.467
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr5_-_24645084
|
1.437
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chrX_-_139866582
|
1.436
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chr11_-_36619776
|
1.436
|
NM_000536
NM_001243785
NM_001243786
|
RAG2
|
recombination activating gene 2
|
|
chr7_+_107110501
|
1.389
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_-_160549226
|
1.375
|
|
CD84
|
CD84 molecule
|
|
chr3_-_150920946
|
1.364
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr14_-_106691698
|
1.360
|
|
|
|
|
chr4_-_25032306
|
1.358
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|