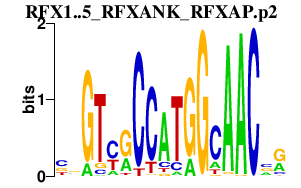
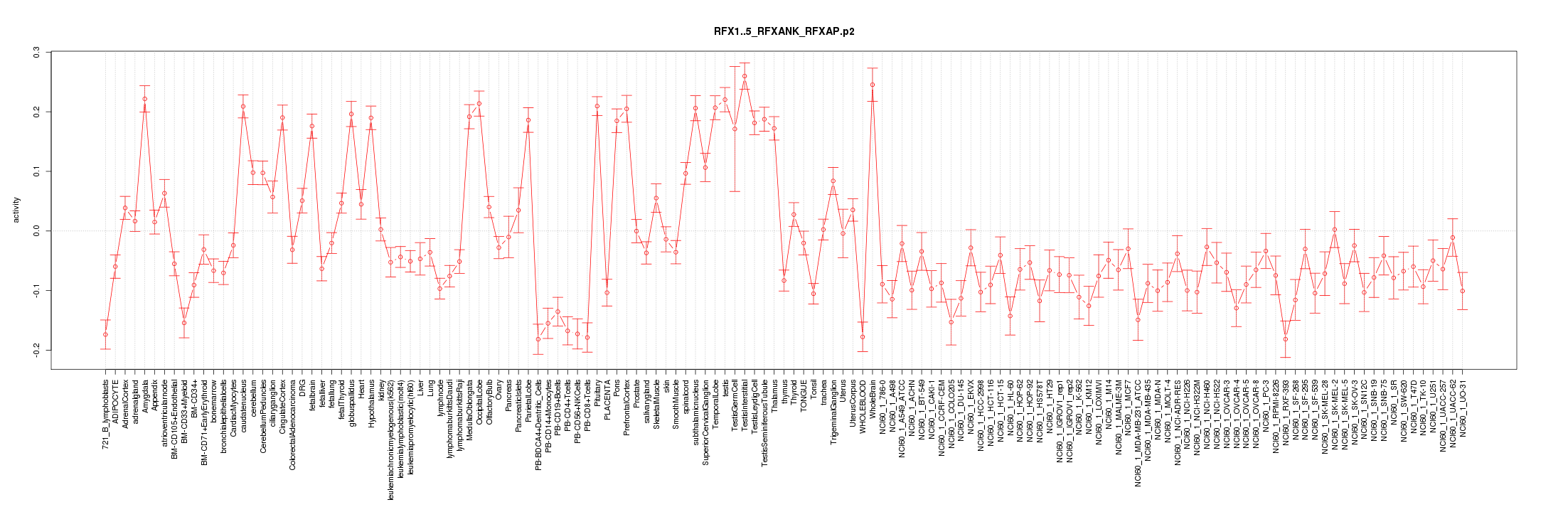
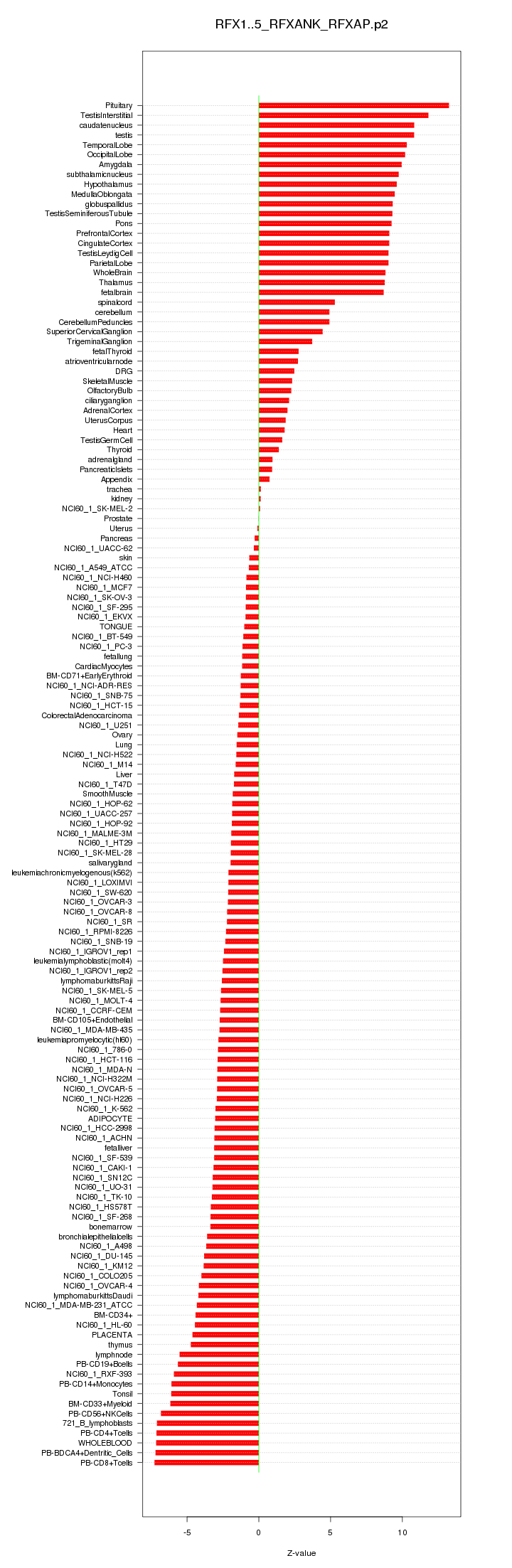
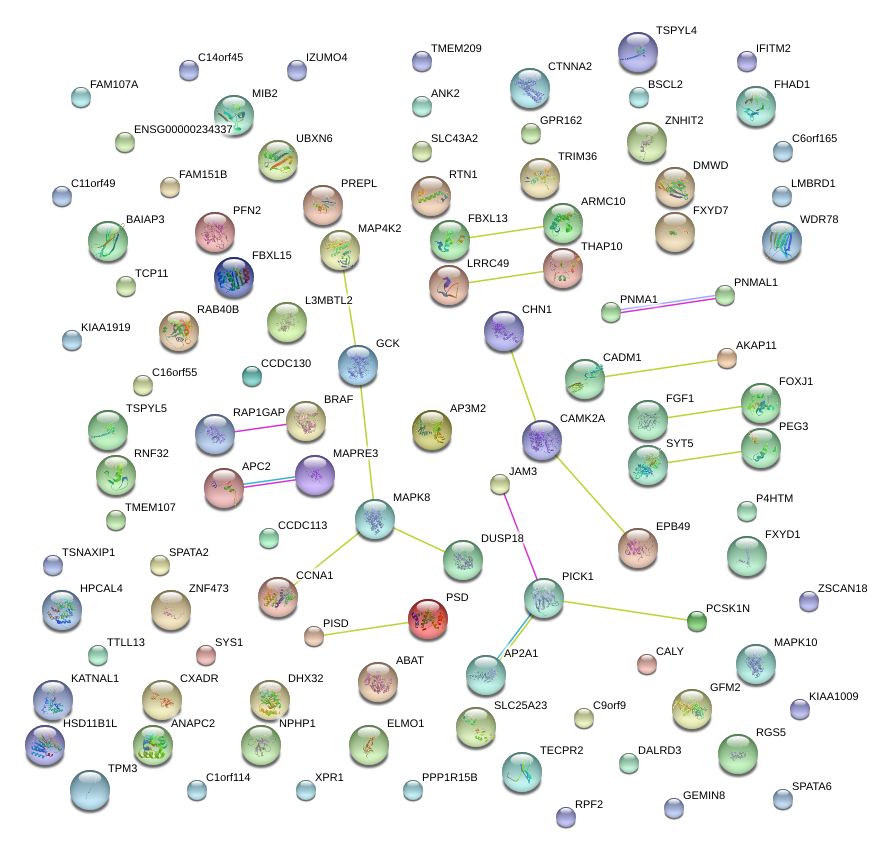
|
chrX_-_48693934
|
120.976
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr11_-_62473502
|
102.367
|
NM_001130702
NM_032667
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr5_-_149669400
|
96.744
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr15_+_71184781
|
84.004
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr14_-_60337350
|
82.734
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr3_-_58563070
|
77.569
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr16_+_8806825
|
74.745
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_+_133938819
|
74.342
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr2_-_175869910
|
72.137
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr15_-_71184611
|
68.310
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr11_+_133938965
|
64.937
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr19_-_55690717
|
64.808
|
|
SYT5
|
synaptotagmin V
|
|
chr14_-_74180341
|
63.828
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chr17_-_5404304
|
58.504
|
NM_001162371
|
LOC728392
|
uncharacterized LOC728392
|
|
chr19_-_46974663
|
57.496
|
|
PNMAL1
|
PNMA-like 1
|
|
chr5_-_149669187
|
56.778
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr17_-_74137370
|
56.355
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr20_+_43991739
|
55.130
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 readthrough
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr6_+_88117689
|
54.858
|
NM_001031743
|
C6orf165
|
chromosome 6 open reading frame 165
|
|
chr1_-_204380903
|
54.855
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr6_+_111303240
|
54.118
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr19_-_58609606
|
52.773
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr19_-_46974789
|
51.603
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr6_-_116575170
|
46.980
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr13_-_30881556
|
46.735
|
NM_032116
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr1_-_154155698
|
45.724
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr19_+_1450117
|
45.043
|
NM_005883
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr2_+_27193448
|
43.731
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr1_-_48937791
|
43.067
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr1_-_48937676
|
42.985
|
|
SPATA6
|
spermatogenesis associated 6
|
|
chr19_-_57352055
|
42.978
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr3_+_49027770
|
42.358
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr1_-_169396654
|
41.473
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr10_-_135150368
|
40.443
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr22_+_38453478
|
40.177
|
|
PICK1
|
protein interacting with PRKCA 1
|
|
chr19_+_35634153
|
39.847
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chr16_+_67840976
|
39.826
|
NM_018430
|
TSNAXIP1
|
translin-associated factor X interacting protein 1
|
|
chr10_-_135150474
|
38.732
|
NM_015722
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr3_+_49027281
|
38.701
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr7_+_102715293
|
36.674
|
NM_001161009
NM_001161010
NM_001161011
NM_001161012
NM_001161013
NM_031905
|
ARMC10
|
armadillo repeat containing 10
|
|
chr5_+_79783799
|
36.524
|
NM_205548
|
FAM151B
|
family with sequence similarity 151, member B
|
|
chr9_+_135754284
|
35.183
|
NM_018956
|
C9orf9
|
chromosome 9 open reading frame 9
|
|
chr1_+_1550794
|
35.078
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr16_+_84178864
|
34.980
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|
|
chr7_-_102715171
|
34.828
|
NM_001111038
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr15_+_90792761
|
34.634
|
NM_001029964
|
TTLL13
|
tubulin tyrosine ligase-like family, member 13
|
|
chr8_-_98290072
|
34.506
|
|
TSPYL5
|
TSPY-like 5
|
|
chr4_-_87281177
|
34.453
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr19_+_2096930
|
34.408
|
|
IZUMO4
|
IZUMO family member 4
|
|
chr6_-_35109003
|
33.850
|
NM_001093728
NM_018679
|
TCP11
|
t-complex 11 homolog (mouse)
|
|
chr17_-_64187814
|
33.808
|
NM_145036
NM_001199165
|
CEP112
|
centrosomal protein 112kDa
|
|
chr19_+_5681016
|
33.769
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr7_-_44228880
|
33.260
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr22_-_31063786
|
33.111
|
NM_152511
|
DUSP18
|
dual specificity phosphatase 18
|
|
chr11_+_46958228
|
32.954
|
NM_001003676
NM_001003677
NM_001003678
NM_024113
|
C11orf49
|
chromosome 11 open reading frame 49
|
|
chr16_+_58283819
|
32.824
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr3_-_149688886
|
32.559
|
|
PFN2
|
profilin 2
|
|
chr17_-_8079607
|
32.383
|
NM_032354
NM_183065
|
TMEM107
|
transmembrane protein 107
|
|
chr7_-_129845216
|
32.024
|
NM_032842
|
TMEM209
|
transmembrane protein 209
|
|
chr7_+_102715580
|
32.018
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr7_-_140624280
|
32.000
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr13_+_37006408
|
31.386
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr11_-_115375065
|
31.371
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr16_+_8814567
|
31.288
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr21_+_18885104
|
31.176
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr17_-_80656461
|
31.169
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr1_-_154155594
|
30.921
|
|
TPM3
|
tropomyosin 3
|
|
chr3_-_49055916
|
30.918
|
NM_001009996
|
DALRD3
|
DALR anticodon binding domain containing 3
|
|
chr16_+_89724175
|
30.682
|
NM_153025
|
C16orf55
|
chromosome 16 open reading frame 55
|
|
chr21_+_18885296
|
30.582
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_-_163113938
|
30.317
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_40157067
|
30.200
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr11_+_67085309
|
30.091
|
|
LOC100130987
|
uncharacterized LOC100130987
|
|
chr19_+_50529294
|
29.752
|
|
ZNF473
|
zinc finger protein 473
|
|
chr5_-_74062885
|
29.751
|
NM_032380
NM_170681
NM_170691
|
GFM2
|
G elongation factor, mitochondrial 2
|
|
chr8_+_21916685
|
29.714
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_+_102829351
|
29.392
|
|
TECPR2
|
tectonin beta-propeller repeat containing 2
|
|
chr8_-_98290171
|
29.153
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr8_+_21916858
|
29.074
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr20_-_48531986
|
28.561
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_+_5681139
|
28.554
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr22_+_41601285
|
28.086
|
NM_031488
|
L3MBTL2
|
l(3)mbt-like 2 (Drosophila)
|
|
chr19_-_4457789
|
27.985
|
NM_025241
|
UBXN6
|
UBX domain protein 6
|
|
chr6_-_70506472
|
27.891
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr6_+_111580481
|
27.730
|
NM_153369
|
KIAA1919
|
KIAA1919
|
|
chr1_-_21978287
|
27.288
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr2_-_44588943
|
27.059
|
NM_001171603
NM_001171617
|
PREPL
|
prolyl endopeptidase-like
|
|
chr7_+_156433352
|
26.913
|
NM_030936
NM_001184996
|
RNF32
|
ring finger protein 32
|
|
chr19_+_50270263
|
26.889
|
|
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit
|
|
chr2_+_79740059
|
26.841
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr13_+_42846255
|
26.613
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr3_-_139108486
|
26.194
|
|
|
|
|
chr5_-_114505549
|
26.109
|
|
TRIM36
|
tripartite motif containing 36
|
|
chr1_+_180601121
|
25.992
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr20_-_48532036
|
25.833
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_+_35630391
|
25.755
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr4_+_113970784
|
25.580
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr10_-_127584967
|
25.176
|
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr5_-_142065991
|
25.090
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_142065586
|
25.087
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_-_84937313
|
25.071
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr8_+_21912327
|
25.060
|
NM_001114136
NM_001114139
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr12_+_6930689
|
25.035
|
NM_014449
NM_019858
|
GPR162
|
G protein-coupled receptor 162
|
|
chrX_-_14047958
|
24.910
|
NM_001042479
NM_001042480
NM_017856
|
GEMIN8
|
gem (nuclear organelle) associated protein 8
|
|
chr8_+_42010463
|
24.635
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr19_+_13842564
|
24.616
|
|
CCDC130
|
coiled-coil domain containing 130
|
|
chr11_-_64885124
|
24.472
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr17_-_1531543
|
24.422
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr2_-_110962577
|
24.277
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr19_-_46295785
|
24.256
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr16_+_1386197
|
24.177
|
NM_001199096
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr11_+_28129797
|
24.152
|
NM_001113528
NM_152636
|
METTL15
|
methyltransferase like 15
|
|
chr2_-_44588622
|
23.908
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr22_+_38453261
|
23.794
|
NM_001039583
NM_001039584
NM_012407
|
PICK1
|
protein interacting with PRKCA 1
|
|
chr1_+_15573767
|
23.514
|
NM_052929
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1
|
|
chr20_-_3869713
|
23.229
|
|
|
|
|
chr20_-_48532067
|
23.192
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr7_-_37025696
|
23.139
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_-_67390413
|
23.111
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr19_+_2096792
|
23.060
|
NM_001031735
NM_001039846
|
IZUMO4
|
IZUMO family member 4
|
|
chr14_+_74486049
|
23.000
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr10_-_104179468
|
22.999
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr8_+_22462286
|
22.890
|
|
KIAA1967
|
KIAA1967
|
|
chr19_+_50529211
|
22.667
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr1_+_38022514
|
22.643
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr6_-_32163890
|
22.537
|
|
NOTCH4
|
notch 4
|
|
chr3_+_130745693
|
22.442
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr10_+_22634373
|
22.424
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr22_-_43485333
|
22.347
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr11_-_118123001
|
22.345
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr5_-_133702764
|
22.192
|
NM_001113575
NM_016508
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr2_+_79740171
|
21.830
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr8_+_22462273
|
21.809
|
|
KIAA1967
|
KIAA1967
|
|
chr10_+_12171580
|
21.781
|
NM_001142627
NM_001142628
NM_018144
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr1_-_85725315
|
21.682
|
NM_198077
|
C1orf52
|
chromosome 1 open reading frame 52
|
|
chr2_+_214149102
|
21.660
|
NM_001025436
NM_024532
|
SPAG16
|
sperm associated antigen 16
|
|
chrX_+_135251795
|
21.596
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_66360485
|
21.547
|
NM_018219
|
CCDC87
|
coiled-coil domain containing 87
|
|
chr13_-_96294282
|
21.415
|
|
DZIP1
|
DAZ interacting protein 1
|
|
chr11_+_66278076
|
21.383
|
NM_024649
|
BBS1
|
Bardet-Biedl syndrome 1
|
|
chr21_+_40817793
|
21.366
|
NM_001001713
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr19_+_50529276
|
21.326
|
|
ZNF473
|
zinc finger protein 473
|
|
chr2_-_2334887
|
21.302
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_72206023
|
21.244
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr19_-_2783313
|
21.134
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr19_-_58874078
|
21.101
|
NM_001207009
NM_198458
|
ZNF497
|
zinc finger protein 497
|
|
chr16_-_72206348
|
21.078
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr1_+_84630374
|
21.071
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr11_-_116968986
|
20.911
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr6_-_121655606
|
20.905
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr8_+_22462267
|
20.830
|
|
KIAA1967
|
KIAA1967
|
|
chr12_-_123459649
|
20.817
|
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr19_-_2783225
|
20.811
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr8_+_22462532
|
20.706
|
NM_021174
|
KIAA1967
|
KIAA1967
|
|
chr19_-_59084735
|
20.678
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr18_+_54318561
|
20.584
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr3_+_184056304
|
20.348
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr14_-_59932051
|
20.285
|
NM_022571
|
GPR135
|
G protein-coupled receptor 135
|
|
chr7_-_17980102
|
20.201
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr16_-_47494753
|
20.201
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr8_+_38243958
|
20.175
|
NM_001199659
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr16_-_19729417
|
20.123
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr17_-_9862765
|
20.114
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_1532109
|
20.105
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr19_-_5680910
|
20.028
|
NM_205767
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr6_+_35227234
|
20.008
|
|
ZNF76
|
zinc finger protein 76
|
|
chr11_-_71791734
|
19.938
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr5_-_157098444
|
19.918
|
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr3_+_50306578
|
19.904
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr13_-_31736031
|
19.839
|
NM_006644
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr1_-_156217717
|
19.807
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr3_-_13114547
|
19.581
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_+_169337193
|
19.491
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_+_38080695
|
19.476
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr19_-_375969
|
19.441
|
NM_016585
NM_199202
|
THEG
|
Theg homolog (mouse)
|
|
chr3_-_113233952
|
19.344
|
NM_144718
|
SPICE1
|
spindle and centriole associated protein 1
|
|
chr9_+_135753711
|
19.200
|
|
C9orf9
|
chromosome 9 open reading frame 9
|
|
chrX_+_135251454
|
19.067
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_-_132073143
|
18.950
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr7_-_4923246
|
18.901
|
NM_018059
|
RADIL
|
Ras association and DIL domains
|
|
chr12_+_2921786
|
18.828
|
NM_018463
|
ITFG2
|
integrin alpha FG-GAP repeat containing 2
|
|
chr16_-_47494981
|
18.763
|
NM_030790
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr19_-_2783332
|
18.679
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr17_-_16472463
|
18.664
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr4_-_156298094
|
18.614
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr13_-_73355969
|
18.583
|
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr8_+_94767071
|
18.515
|
NM_001142301
NM_153704
|
TMEM67
|
transmembrane protein 67
|
|
chr3_-_196439067
|
18.507
|
NM_032898
|
CEP19
|
centrosomal protein 19kDa
|
|
chr19_+_50879681
|
18.455
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr5_-_147162175
|
18.440
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr8_-_110704019
|
18.304
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr13_-_44361032
|
18.152
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr15_-_55700456
|
18.136
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr19_+_56652529
|
17.996
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr3_+_196439211
|
17.859
|
NM_001166304
NM_017861
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X
|
|
chr4_-_156297944
|
17.802
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr2_+_173600595
|
17.802
|
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr17_-_1012257
|
17.780
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr10_-_104178900
|
17.549
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr2_+_230787195
|
17.545
|
NM_174899
|
FBXO36
|
F-box protein 36
|