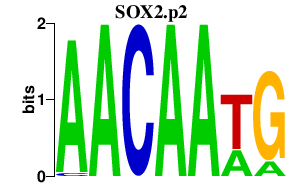
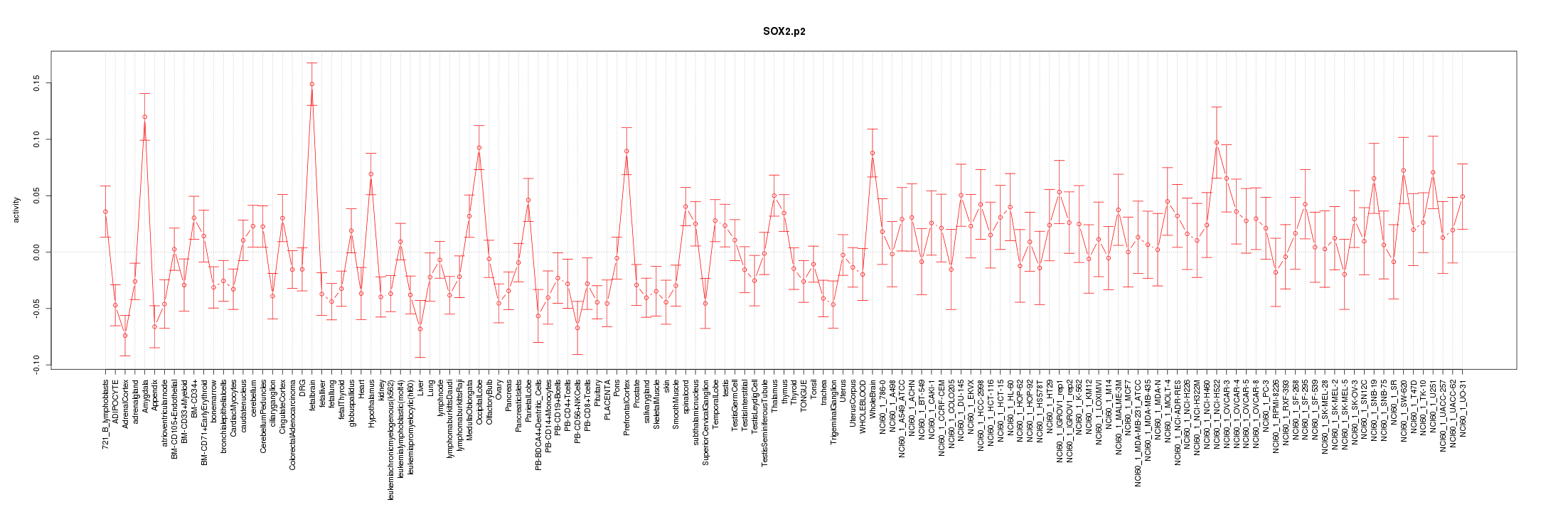
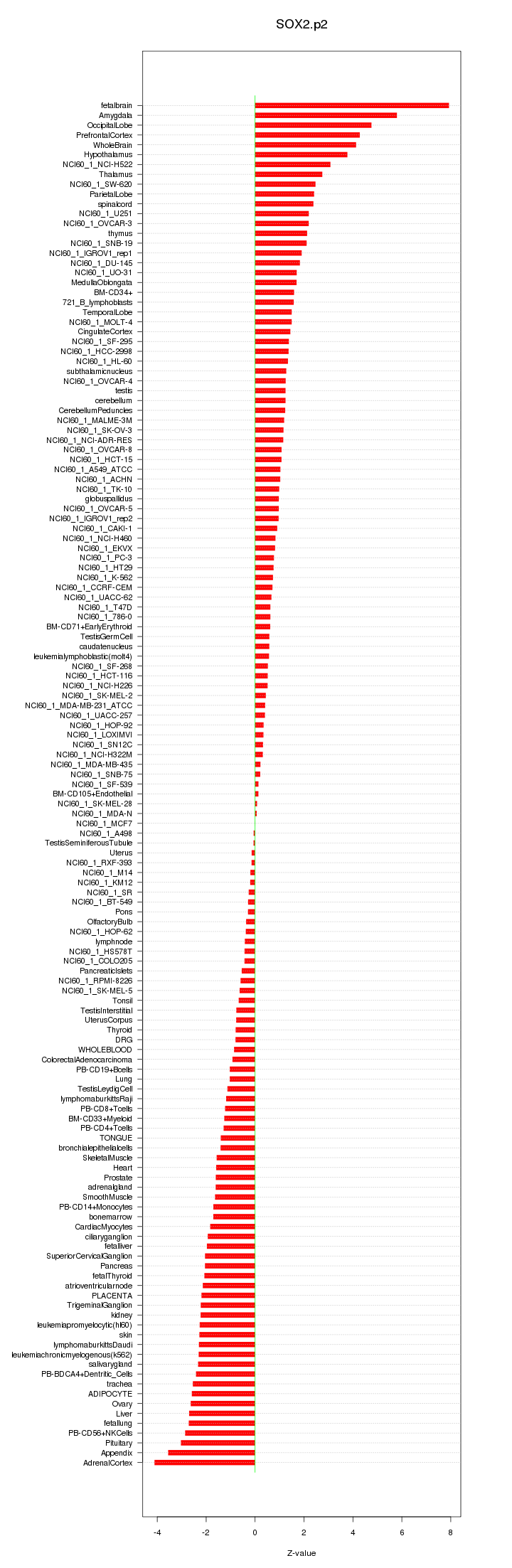
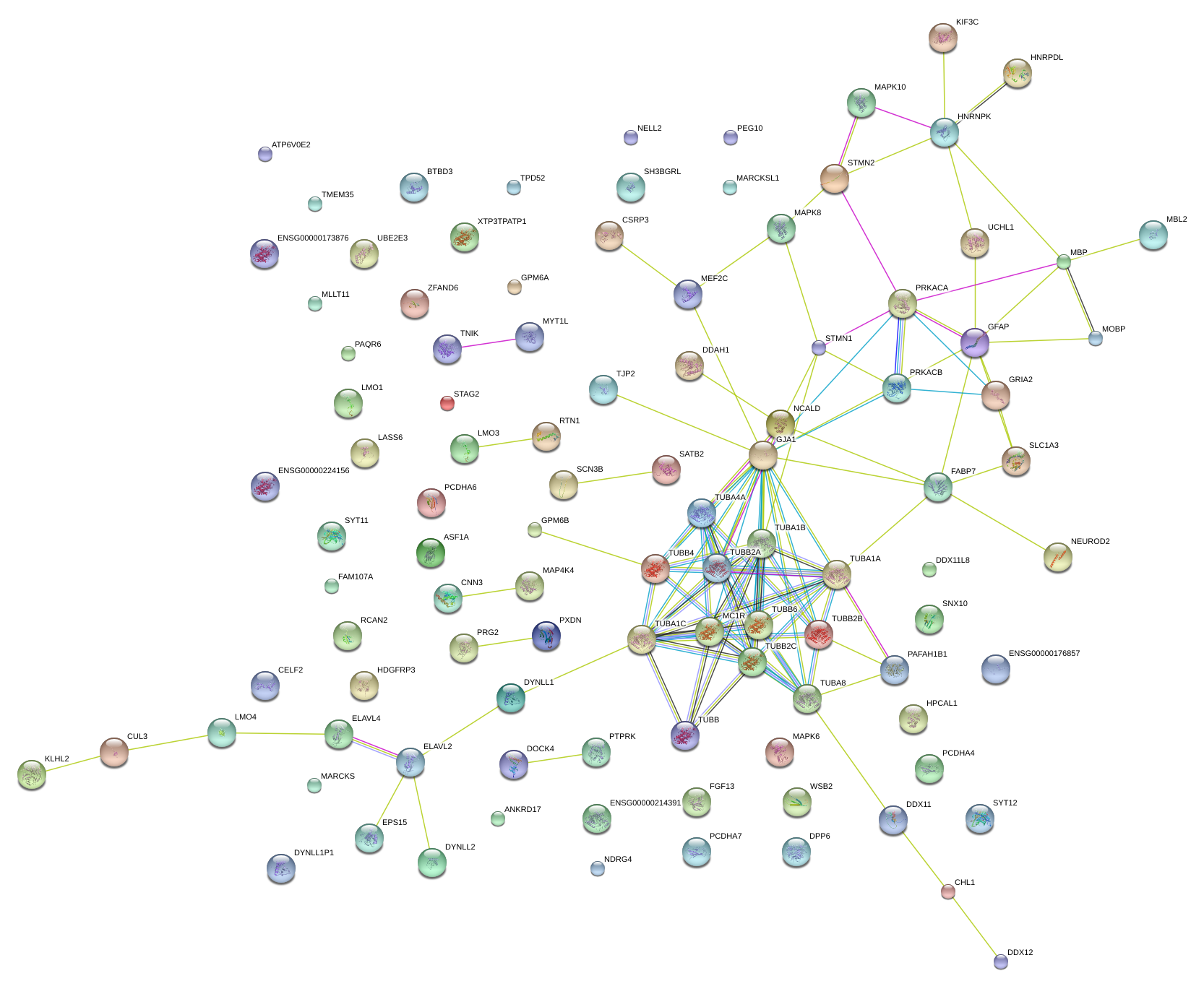
|
chr1_+_84647342
|
38.977
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr20_+_11898536
|
38.923
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr4_-_176733413
|
36.198
|
|
GPM6A
|
glycoprotein M6A
|
|
chr3_+_39509069
|
32.924
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr2_+_102508333
|
30.470
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr1_-_32801606
|
29.340
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr2_-_2334887
|
26.738
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr7_+_26400503
|
25.897
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chrX_+_123234462
|
25.820
|
|
STAG2
|
stromal antigen 2
|
|
chr1_-_26233367
|
23.670
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr15_+_52311408
|
23.243
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr2_+_102508377
|
23.123
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr12_-_45307710
|
20.796
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr4_+_166131170
|
20.462
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr15_+_52311438
|
20.187
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr15_+_52311432
|
20.038
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr4_+_41258941
|
19.787
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr7_+_94297448
|
19.575
|
|
PEG10
|
paternally expressed 10
|
|
chr4_-_176733901
|
19.505
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_50571963
|
19.158
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr16_+_89989686
|
18.631
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr6_+_123100645
|
18.158
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr14_-_60097223
|
17.994
|
|
RTN1
|
reticulon 1
|
|
chr3_-_58563490
|
17.471
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr6_+_114178516
|
17.373
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr2_-_55199894
|
17.008
|
|
|
|
|
chr1_+_87797371
|
16.723
|
|
LMO4
|
LIM domain only 4
|
|
chr9_-_86584079
|
16.707
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr7_+_149570004
|
16.697
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr3_-_58563070
|
16.599
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr1_+_151032866
|
16.348
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr12_-_16759531
|
16.178
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr18_-_74728963
|
16.161
|
|
MBP
|
myelin basic protein
|
|
chr1_+_151032880
|
16.148
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr18_-_74729049
|
16.088
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr4_+_41258909
|
15.942
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_41258895
|
15.836
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr17_-_1248346
|
15.827
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr15_+_80364902
|
15.303
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr12_-_16759430
|
15.245
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_111846384
|
15.170
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr17_-_42992852
|
15.147
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr1_+_151032683
|
15.130
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_85930610
|
15.098
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_-_83347222
|
14.852
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr19_-_6502224
|
14.795
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr16_+_89988416
|
14.692
|
NM_001197181
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr8_+_80523351
|
14.556
|
|
STMN2
|
stathmin-like 2
|
|
chr12_-_16757944
|
14.540
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr16_+_58534045
|
14.486
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr2_+_181845605
|
14.455
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr2_-_26205437
|
14.232
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr2_-_26205327
|
13.745
|
|
KIF3C
|
kinesin family member 3C
|
|
chr8_+_80523048
|
13.715
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr1_-_95392501
|
13.431
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr5_+_140213968
|
13.369
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr8_+_80523376
|
13.324
|
|
STMN2
|
stathmin-like 2
|
|
chr12_+_120933911
|
13.315
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr5_+_36606667
|
13.157
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr15_-_83876769
|
13.072
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chrX_+_80457302
|
13.065
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr14_-_60097314
|
13.050
|
|
RTN1
|
reticulon 1
|
|
chr1_-_86043932
|
12.956
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_121756744
|
12.837
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chrX_-_13835187
|
12.795
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_36606456
|
12.771
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr2_+_181845803
|
12.761
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chrX_-_13835012
|
12.608
|
|
GPM6B
|
glycoprotein M6B
|
|
chr12_-_118498893
|
12.582
|
NM_018639
|
WSB2
|
WD repeat and SOCS box containing 2
|
|
chr8_+_80523304
|
12.486
|
|
STMN2
|
stathmin-like 2
|
|
chr3_+_238274
|
12.461
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr5_+_140739589
|
12.186
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr8_+_80523339
|
12.130
|
|
STMN2
|
stathmin-like 2
|
|
chr9_+_71819926
|
12.055
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr1_+_155853546
|
11.953
|
|
SYT11
|
synaptotagmin XI
|
|
chr12_-_49582800
|
11.930
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr2_+_149633173
|
11.913
|
|
|
|
|
chr1_-_51887740
|
11.827
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr1_+_50574601
|
11.738
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_37764165
|
11.702
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr5_+_140254930
|
11.686
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr1_+_50574589
|
11.580
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_74088773
|
11.572
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr2_-_200325200
|
11.541
|
|
SATB2
|
SATB homeobox 2
|
|
chr5_+_140186658
|
11.525
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr6_-_128841432
|
11.396
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr12_+_120933832
|
11.385
|
NM_001037495
NM_003746
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr16_+_58497548
|
11.361
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr10_+_11207053
|
11.242
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr16_+_58497628
|
11.236
|
|
NDRG4
|
NDRG family member 4
|
|
chr8_-_80993009
|
11.235
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr6_-_46293404
|
10.997
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_-_13835313
|
10.915
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr2_+_169312793
|
10.848
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr6_+_119215240
|
10.612
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chrX_+_100333835
|
10.517
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chrX_-_138287180
|
10.485
|
NM_001139498
NM_001139500
NM_001139501
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_95392449
|
10.476
|
|
CNN3
|
calponin 3, acidic
|
|
chr8_-_102803178
|
10.334
|
|
NCALD
|
neurocalcin delta
|
|
chrX_+_80457597
|
10.321
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_-_13835213
|
10.293
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_156217717
|
10.282
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr4_+_158141735
|
10.222
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr2_-_225370687
|
10.208
|
|
CUL3
|
cullin 3
|
|
chr4_-_87028805
|
10.198
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr8_-_102803196
|
10.194
|
|
NCALD
|
neurocalcin delta
|
|
chr11_-_123524881
|
10.041
|
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr14_-_60097531
|
10.030
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr6_+_123100860
|
9.996
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr5_-_88199868
|
9.964
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_+_154002194
|
9.860
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr6_+_30688085
|
9.752
|
|
TUBB
|
tubulin, beta class I
|
|
chr11_-_117747331
|
9.722
|
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr17_-_29624249
|
9.627
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr15_-_64995346
|
9.616
|
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr15_+_91643181
|
9.587
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_18666295
|
9.571
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_+_49212253
|
9.554
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr4_-_176734181
|
9.523
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr1_-_149900143
|
9.505
|
NM_005850
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa
|
|
chr15_+_84115979
|
9.474
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr10_+_11206928
|
9.412
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr22_-_50523853
|
9.408
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr1_+_87794576
|
9.388
|
|
LMO4
|
LIM domain only 4
|
|
chr12_-_16758839
|
9.287
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_32388687
|
9.152
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr13_-_24007822
|
9.103
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr1_+_244216506
|
9.086
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr5_+_71502700
|
9.039
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr8_+_26435920
|
9.014
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr2_+_102314162
|
8.971
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr13_-_53422773
|
8.945
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr12_+_69979447
|
8.841
|
NM_001198842
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr1_+_163038564
|
8.840
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_+_36608430
|
8.819
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_+_163038934
|
8.791
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_-_111091947
|
8.791
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr21_-_32931289
|
8.783
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr4_-_87281374
|
8.540
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr5_+_140749830
|
8.522
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr2_-_55277592
|
8.509
|
|
RTN4
|
reticulon 4
|
|
chr19_-_42806460
|
8.444
|
NM_002573
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa)
|
|
chr4_+_41614911
|
8.425
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_-_10830581
|
8.383
|
NM_001042559
NM_001418
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr19_-_42806928
|
8.368
|
NM_001145939
NM_001145940
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa)
|
|
chr12_+_120933916
|
8.344
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr5_+_137673223
|
8.313
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr2_+_10262853
|
8.312
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr2_-_55277421
|
8.294
|
|
RTN4
|
reticulon 4
|
|
chr4_-_83347778
|
8.218
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr5_-_146435589
|
8.187
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_+_244214576
|
8.180
|
|
ZNF238
|
zinc finger protein 238
|
|
chr4_-_83350548
|
8.178
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr2_-_74601653
|
8.175
|
NM_001135041
NM_023019
|
DCTN1
|
dynactin 1
|
|
chr5_+_65372744
|
8.140
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr15_+_57210972
|
8.130
|
|
TCF12
|
transcription factor 12
|
|
chrX_+_51927918
|
8.005
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr2_-_55277672
|
7.937
|
|
RTN4
|
reticulon 4
|
|
chr18_-_21852189
|
7.935
|
NM_018030
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr5_-_11903964
|
7.926
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr5_-_32388595
|
7.898
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr2_+_10262694
|
7.891
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr15_+_65822842
|
7.883
|
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr1_-_110283659
|
7.873
|
NM_000849
|
GSTM3
|
glutathione S-transferase mu 3 (brain)
|
|
chr2_-_55277620
|
7.822
|
|
RTN4
|
reticulon 4
|
|
chr2_-_55277653
|
7.813
|
|
RTN4
|
reticulon 4
|
|
chr21_+_18885104
|
7.782
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr5_+_140797243
|
7.771
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr4_+_113970784
|
7.712
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr7_-_47621635
|
7.709
|
|
TNS3
|
tensin 3
|
|
chr5_-_16936371
|
7.676
|
NM_012334
|
MYO10
|
myosin X
|
|
chr15_+_22892683
|
7.674
|
NM_014608
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr11_-_72380107
|
7.664
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr5_+_65222381
|
7.615
|
NM_001006600
NM_001253697
NM_001253698
NM_001253699
NM_001253701
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chrX_-_51812249
|
7.586
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr11_-_117166247
|
7.558
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr2_-_175869910
|
7.543
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr2_+_62934030
|
7.524
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr6_+_30688144
|
7.515
|
NM_178014
|
TUBB
|
tubulin, beta class I
|
|
chr4_-_76598289
|
7.484
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr5_+_140797270
|
7.473
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr2_-_55277698
|
7.469
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr18_+_29171729
|
7.442
|
NM_000371
|
TTR
|
transthyretin
|
|
chr5_-_146460991
|
7.437
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr11_-_10830452
|
7.371
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr2_-_55277459
|
7.342
|
|
RTN4
|
reticulon 4
|
|
chr6_-_75965845
|
7.320
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr6_-_79944397
|
7.314
|
NM_001201362
NM_001201363
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr6_-_79944324
|
7.253
|
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr3_+_159557745
|
7.238
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr10_-_62332666
|
7.220
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr15_+_22892714
|
7.189
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr7_-_47621741
|
7.187
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr1_-_177133950
|
7.141
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr12_-_45270071
|
7.140
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr2_-_166060552
|
7.136
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr15_+_22892699
|
7.126
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr16_+_6069094
|
7.113
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr22_-_50523737
|
7.103
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr15_+_22892723
|
7.095
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|