|
chr20_+_35169876
|
210.156
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr20_+_35169897
|
178.254
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr2_+_74120132
|
132.470
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr2_+_74120092
|
129.384
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr11_+_117070039
|
111.337
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr5_+_137801178
|
110.076
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr5_+_137801166
|
103.253
|
|
EGR1
|
early growth response 1
|
|
chr10_-_90712510
|
82.201
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr3_-_123339423
|
62.395
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_123339178
|
60.160
|
|
MYLK
|
myosin light chain kinase
|
|
chr19_-_45826131
|
56.558
|
|
CKM
|
creatine kinase, muscle
|
|
chr1_-_229569839
|
50.036
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr19_-_45826232
|
48.687
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr15_-_35087748
|
36.898
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr19_-_36247917
|
36.320
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr10_-_29923900
|
34.555
|
NM_021738
|
SVIL
|
supervillin
|
|
chr22_+_31480981
|
34.248
|
NM_001207018
|
SMTN
|
smoothelin
|
|
chr7_-_107643330
|
30.485
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr4_-_88450601
|
28.613
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr6_+_151561094
|
27.144
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr10_+_75757847
|
27.010
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr2_-_106015423
|
26.379
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr12_-_50616425
|
26.301
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr7_-_94285426
|
25.779
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr7_-_94285477
|
25.611
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr16_-_11375094
|
25.509
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr7_-_94285372
|
25.021
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr15_+_96869156
|
24.509
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_-_99594915
|
24.148
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr14_-_53417808
|
23.817
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr20_+_4702499
|
23.343
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr12_+_52445185
|
21.946
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr7_+_143078399
|
21.380
|
|
ZYX
|
zyxin
|
|
chr7_+_143078358
|
20.940
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chrX_-_153602928
|
20.924
|
|
FLNA
|
filamin A, alpha
|
|
chr11_+_75273245
|
20.692
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr1_+_223889253
|
19.868
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr7_+_134464161
|
19.700
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr1_-_16344438
|
19.644
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr3_-_46904879
|
19.370
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr17_+_42634598
|
19.317
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr6_+_151561508
|
19.058
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_+_11994710
|
18.619
|
NM_000302
|
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr1_-_20306093
|
18.429
|
NM_001161729
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr7_+_143078444
|
18.293
|
|
ZYX
|
zyxin
|
|
chr16_+_56966025
|
17.853
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr1_-_89530890
|
17.845
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr7_+_143078395
|
17.548
|
|
ZYX
|
zyxin
|
|
chr3_+_187930720
|
17.491
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_143078447
|
17.349
|
|
ZYX
|
zyxin
|
|
chr15_+_96873845
|
16.908
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr7_+_143078457
|
16.000
|
|
ZYX
|
zyxin
|
|
chr19_+_45971249
|
15.778
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chrX_-_153602998
|
15.505
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr7_-_27183225
|
15.179
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr11_-_65667860
|
14.133
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr17_+_41003184
|
13.961
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr4_+_24797084
|
13.909
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr15_+_74218962
|
13.875
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr4_-_88450243
|
13.520
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr3_+_187871473
|
13.321
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr11_-_65667858
|
13.222
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr14_+_75745480
|
12.732
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr12_-_91505541
|
12.705
|
NM_002345
|
LUM
|
lumican
|
|
chr10_-_16859389
|
12.414
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr2_-_128432642
|
12.320
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr11_-_65667809
|
12.055
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_16178170
|
11.960
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr9_+_116356199
|
11.874
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr12_-_11508509
|
11.705
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chr15_-_65360297
|
11.658
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr2_+_26915390
|
11.606
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr17_-_79479780
|
11.370
|
|
ACTG1
|
actin, gamma 1
|
|
chr16_+_31483066
|
10.437
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr12_-_11548485
|
9.681
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr7_+_143078608
|
9.654
|
|
ZYX
|
zyxin
|
|
chr17_-_79479831
|
9.582
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr18_-_3219846
|
9.246
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr19_+_2476122
|
9.166
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr1_-_154164551
|
9.092
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chrX_-_11284094
|
8.804
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chrX_-_153599719
|
8.721
|
|
FLNA
|
filamin A, alpha
|
|
chr10_+_112257624
|
8.641
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr4_-_10118460
|
8.605
|
|
WDR1
|
WD repeat domain 1
|
|
chr4_-_10118444
|
8.374
|
|
WDR1
|
WD repeat domain 1
|
|
chr11_+_66314373
|
8.158
|
NM_001104
|
ACTN3
|
actinin, alpha 3
|
|
chr17_-_46671043
|
8.049
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr2_+_139259476
|
8.037
|
|
|
|
|
chr1_+_11994778
|
8.015
|
|
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr12_-_11463353
|
7.885
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr2_-_151344145
|
7.869
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr7_+_93551015
|
7.470
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr8_-_48650683
|
7.135
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_-_208084432
|
6.937
|
NM_001025109
NM_001773
|
CD34
|
CD34 molecule
|
|
chr5_-_33891967
|
6.810
|
NM_030955
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr6_+_43139190
|
6.808
|
|
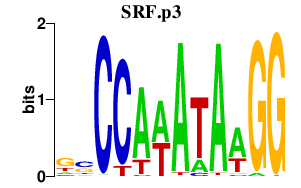
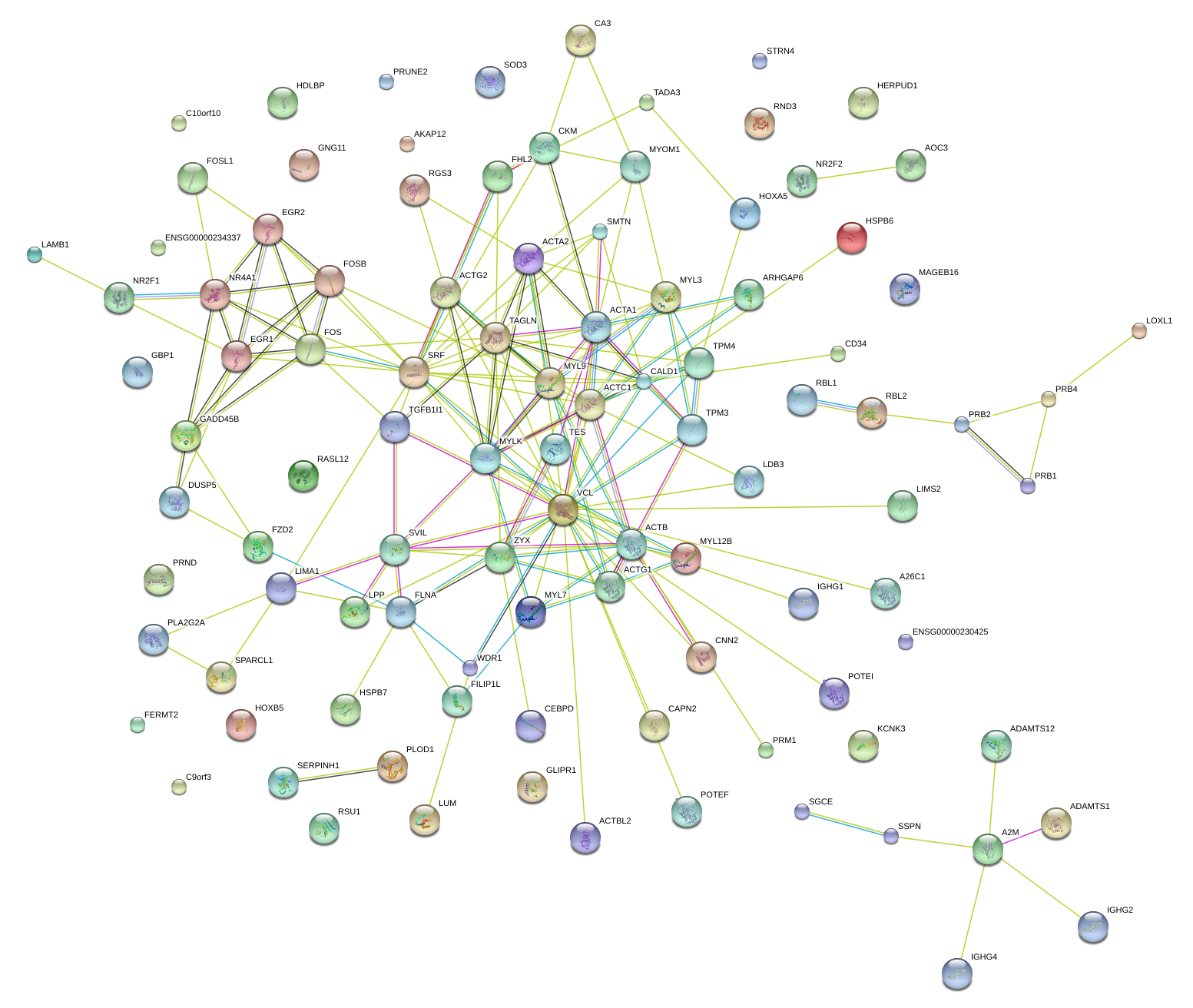
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr10_-_45474210
|
6.781
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr21_-_28217272
|
6.758
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr12_-_9268522
|
6.662
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr9_-_79520878
|
6.610
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr14_-_106111114
|
6.587
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr4_-_10118500
|
6.450
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|
|
chr12_+_75874512
|
6.414
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr9_+_116356407
|
6.335
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_97521930
|
6.265
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr17_-_46671322
|
6.231
|
|
HOXB5
|
homeobox B5
|
|
chrX_+_35816458
|
5.983
|
NM_001099921
|
MAGEB16
|
melanoma antigen family B, 16
|
|
chr19_+_16187306
|
5.958
|
|
TPM4
|
tropomyosin 4
|
|
chr5_-_33892292
|
5.901
|
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr19_+_1026627
|
5.862
|
|
CNN2
|
calponin 2
|
|
chr10_+_88428167
|
5.820
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr10_-_64576074
|
5.746
|
|
EGR2
|
early growth response 2
|
|
chr12_+_26348488
|
5.595
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr2_-_242254984
|
5.579
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr18_-_3219986
|
5.378
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr7_+_115863004
|
5.361
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr8_+_86351055
|
5.123
|
NM_005181
|
CA3
|
carbonic anhydrase III, muscle specific
|
|
chr7_-_44180911
|
4.923
|
NM_021223
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr6_+_43139201
|
4.853
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr3_-_9834357
|
4.779
|
NM_006354
NM_133480
|
TADA3
|
transcriptional adaptor 3
|
|
chr17_-_39684585
|
4.658
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr14_-_106209380
|
4.407
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_106092256
|
4.320
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr10_+_88428263
|
4.264
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr1_+_244816434
|
4.260
|
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr1_-_242162364
|
4.241
|
NM_001004343
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma
|
|
chr22_-_36784055
|
4.214
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr17_+_80193649
|
4.160
|
NM_001206952
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr19_+_57901188
|
4.141
|
NM_001172773
NM_152909
|
ZNF548
|
zinc finger protein 548
|
|
chr5_+_140749830
|
4.116
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr15_-_45406345
|
4.100
|
NM_014080
|
DUOX2
|
dual oxidase 2
|
|
chr1_-_168106535
|
3.937
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr17_+_32612686
|
3.731
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr10_+_123922940
|
3.690
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr19_+_16187047
|
3.392
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr19_+_1026608
|
3.296
|
|
CNN2
|
calponin 2
|
|
chr5_-_39425297
|
3.247
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chrX_-_10851808
|
3.244
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr15_-_45406322
|
3.243
|
|
DUOX2
|
dual oxidase 2
|
|
chrX_-_32430370
|
3.197
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr1_+_19970674
|
3.164
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr14_-_23877480
|
3.116
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr7_-_5569200
|
3.097
|
|
ACTB
|
actin, beta
|
|
chr7_-_80548666
|
3.053
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr1_-_11907741
|
2.975
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr6_-_29527701
|
2.929
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr1_-_35324645
|
2.916
|
NM_001164824
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr1_-_168106810
|
2.855
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr17_-_73505914
|
2.841
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr4_+_87968191
|
2.830
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr11_-_19223516
|
2.827
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chrX_+_99839789
|
2.761
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr7_-_80548322
|
2.724
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr1_-_89591685
|
2.696
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr19_+_45417576
|
2.665
|
|
APOC1
|
apolipoprotein C-I
|
|
chr10_+_31610063
|
2.653
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr17_-_1389013
|
2.597
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr6_+_144612872
|
2.557
|
NM_007124
|
UTRN
|
utrophin
|
|
chr1_-_168106727
|
2.545
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr1_-_150552077
|
2.485
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr20_-_62258330
|
2.451
|
NM_012384
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr10_-_16859336
|
2.354
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr19_+_1026271
|
2.338
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr6_-_73935174
|
2.281
|
NM_001126063
|
KHDC1L
|
KH homology domain containing 1-like
|
|
chr2_-_161056762
|
2.249
|
|
ITGB6
|
integrin, beta 6
|
|
chr6_-_29527477
|
2.223
|
|
UBD
|
ubiquitin D
|
|
chr18_+_3247527
|
2.198
|
NM_006471
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr1_-_150552005
|
2.161
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr11_-_30605748
|
2.112
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr5_-_169816637
|
2.006
|
NM_004137
|
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1
|
|
chr10_-_16859408
|
1.987
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr12_+_50497790
|
1.931
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr6_+_25754926
|
1.847
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr1_+_43148048
|
1.815
|
NM_004559
|
YBX1
|
Y box binding protein 1
|
|
chr18_-_44550533
|
1.803
|
NM_001242907
NM_145653
|
LOC100506888
TCEB3C
|
transcription elongation factor B polypeptide 3C-like
transcription elongation factor B polypeptide 3C (elongin A3)
|
|
chr1_-_31845818
|
1.787
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr4_-_185747051
|
1.772
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr7_-_559479
|
1.754
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr2_-_161056573
|
1.724
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr11_-_75921785
|
1.713
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr14_-_21944826
|
1.663
|
NM_001163380
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr11_+_58390145
|
1.640
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr19_-_46283763
|
1.637
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr3_+_25470067
|
1.636
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr10_+_88516340
|
1.583
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr17_-_66287256
|
1.560
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr8_-_62602285
|
1.461
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr20_-_62680880
|
1.430
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chrX_-_45060081
|
1.387
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr1_+_167691156
|
1.384
|
NM_001146191
NM_003953
NM_024569
|
MPZL1
|
myelin protein zero-like 1
|
|
chr18_-_47806361
|
1.359
|
NM_001204151
|
MBD1
|
methyl-CpG binding domain protein 1
|
|
chr1_+_153388999
|
1.342
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr1_+_43996478
|
1.338
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr10_+_6186758
|
1.314
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr10_+_106034886
|
1.290
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr15_-_59665053
|
1.190
|
|
MYO1E
|
myosin IE
|
|
chr15_-_59665069
|
1.149
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr7_+_12726413
|
1.143
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr19_+_45417920
|
1.142
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr21_+_35190586
|
1.128
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|