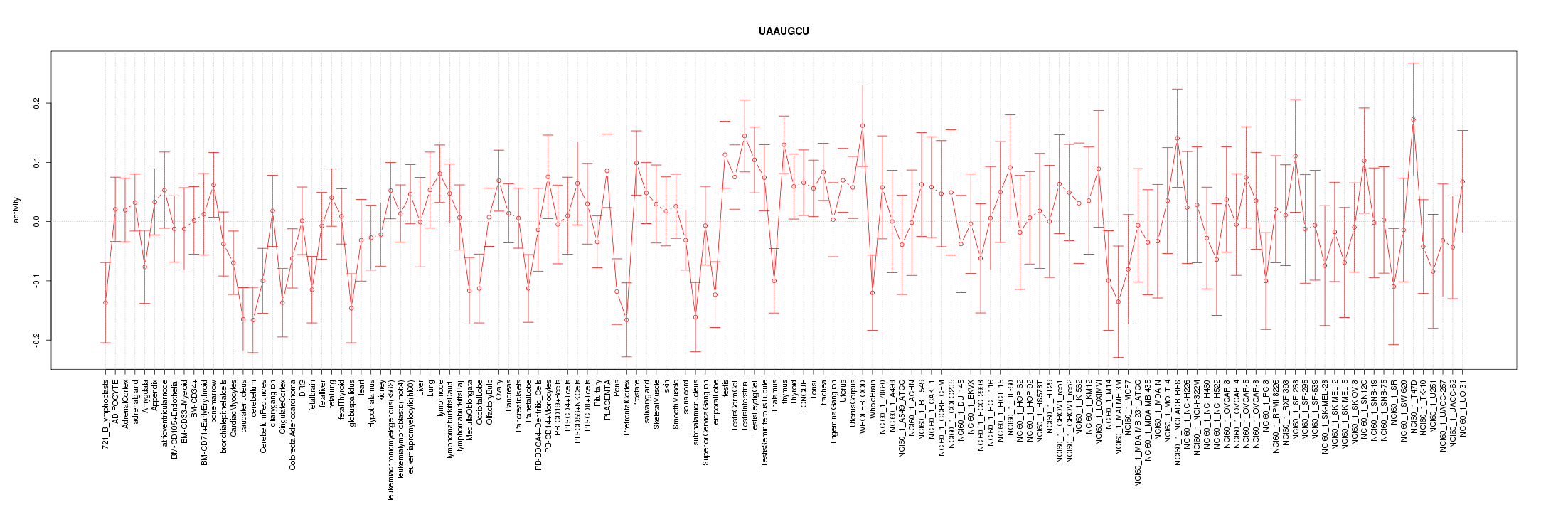
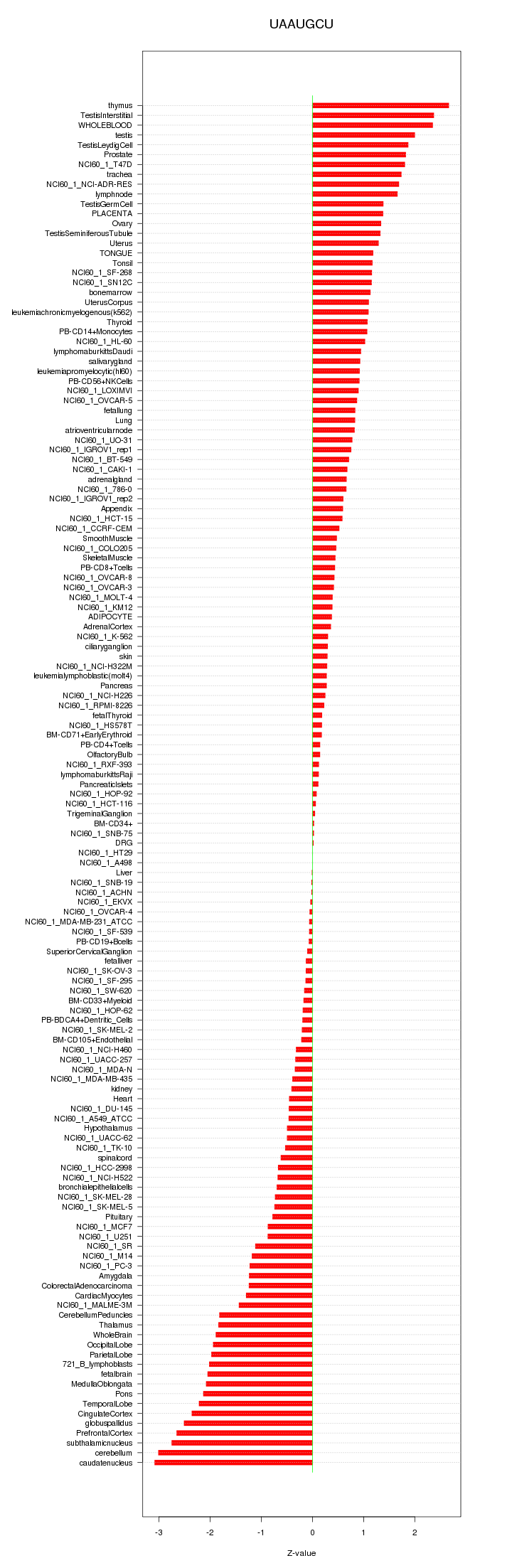
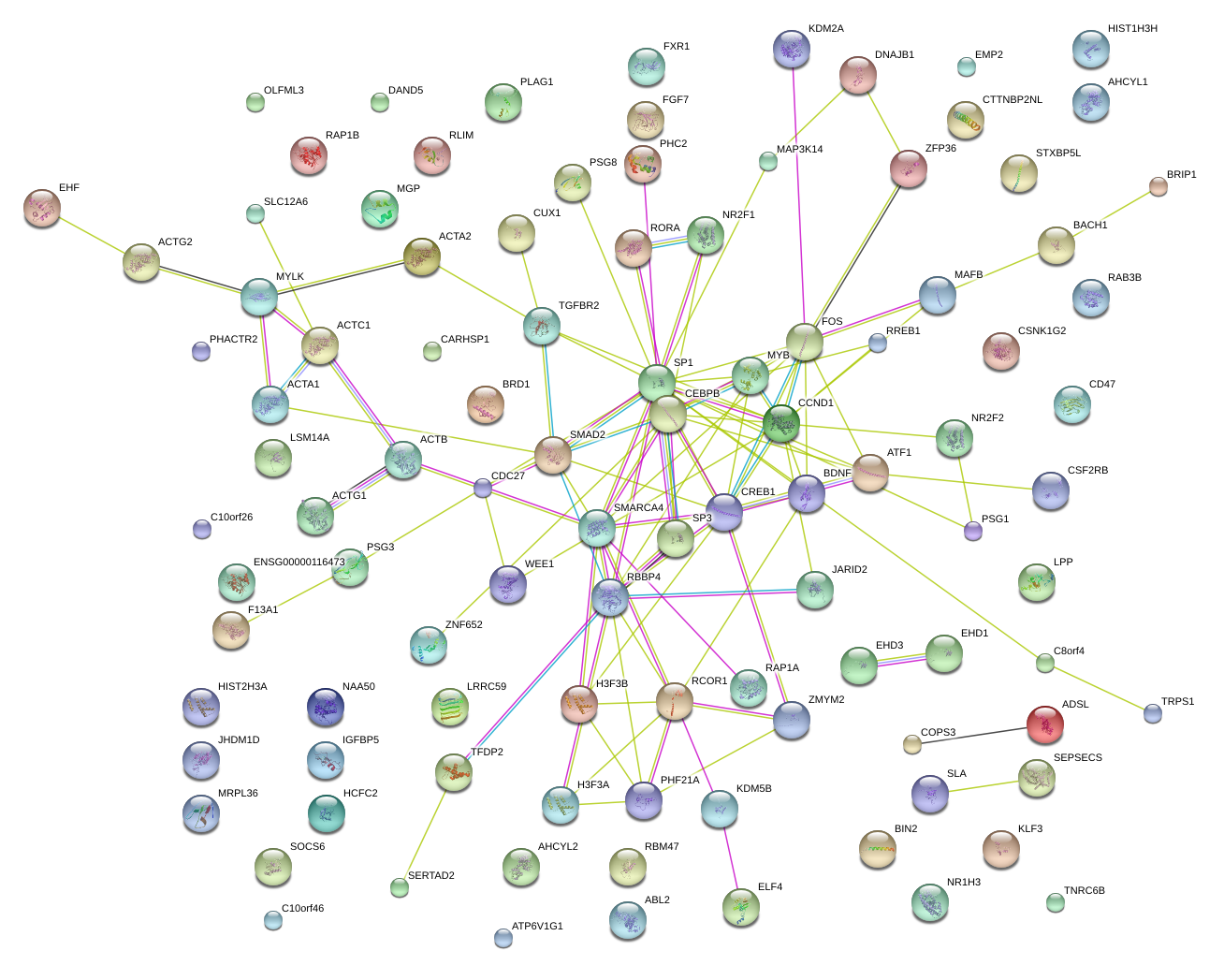
|
chr12_+_69004614
|
11.735
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr20_+_48807119
|
11.028
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chrX_-_73834460
|
9.848
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr4_+_38665587
|
9.099
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_-_141747434
|
8.609
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr14_+_75745480
|
8.515
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr6_+_7107986
|
8.451
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr20_-_39317875
|
8.254
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_-_202777545
|
7.991
|
NM_006618
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr3_-_141868208
|
7.630
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr19_+_39897457
|
7.541
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr3_-_123339423
|
7.375
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_15038724
|
7.103
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr10_+_104535887
|
7.066
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_-_141719188
|
6.986
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr17_-_47439326
|
6.941
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr7_-_139876718
|
6.895
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr15_+_96869156
|
6.847
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr10_-_120514669
|
6.215
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr2_-_64881040
|
6.183
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr14_+_103058988
|
6.124
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr3_-_123603144
|
6.065
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr11_-_64646053
|
6.042
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr15_+_96876568
|
6.006
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr18_-_45457454
|
5.838
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr18_-_45456925
|
5.655
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr15_+_96875793
|
5.651
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_+_96873845
|
5.590
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_+_7107799
|
5.353
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr8_-_116681103
|
4.955
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr11_-_27742224
|
4.881
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr22_+_37309618
|
4.818
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr11_+_9596233
|
4.798
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr16_-_8962842
|
4.794
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr6_+_15246299
|
4.770
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr16_-_8962247
|
4.713
|
NM_001042476
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr10_+_104503724
|
4.640
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr16_-_10674443
|
4.598
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr19_-_14629160
|
4.314
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr4_-_40631846
|
4.300
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr3_-_107809754
|
4.271
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr3_+_187871473
|
4.171
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_+_1941157
|
4.168
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr3_-_113465101
|
4.091
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr11_+_69455837
|
4.028
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr1_-_33841142
|
3.842
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr1_+_226250407
|
3.791
|
NM_002107
|
H3F3A
|
H3 histone, family 3A
|
|
chr6_-_6320897
|
3.749
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr11_-_27741192
|
3.690
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_+_53774422
|
3.657
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr18_+_67956136
|
3.622
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr3_+_187943192
|
3.610
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_51717905
|
3.596
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr7_+_101460881
|
3.542
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr4_-_40517913
|
3.539
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_229569839
|
3.536
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chrX_-_129244652
|
3.530
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr12_+_53773924
|
3.359
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chrX_-_129244471
|
3.337
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr8_-_134115309
|
3.294
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr11_+_34654010
|
3.261
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr9_+_117349993
|
3.197
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr11_-_46142955
|
3.194
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr11_+_9595183
|
3.192
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr1_-_52456292
|
3.018
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr2_-_174828775
|
3.005
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr3_+_30647901
|
3.002
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr3_+_180630054
|
2.968
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr11_-_27723061
|
2.955
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr19_+_34663311
|
2.896
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr15_-_34629941
|
2.844
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_-_179198814
|
2.757
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr17_-_17184589
|
2.745
|
NM_003653
|
COPS3
|
COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis)
|
|
chr2_-_174830429
|
2.734
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr8_-_57123858
|
2.716
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr15_-_60884615
|
2.707
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_34610928
|
2.705
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_33116748
|
2.701
|
NM_001135255
NM_005610
NM_001135256
|
RBBP4
|
retinoblastoma binding protein 4
|
|
chr15_+_49715374
|
2.699
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr3_+_120627026
|
2.681
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr19_+_11071583
|
2.644
|
NM_001128844
NM_003072
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr22_+_40573879
|
2.615
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_187930720
|
2.614
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr22_-_50218451
|
2.594
|
NM_014577
|
BRD1
|
bromodomain containing 1
|
|
chr11_+_66886735
|
2.587
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr6_+_135502445
|
2.587
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr2_-_217560247
|
2.551
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr1_+_114522029
|
2.544
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr11_+_47279467
|
2.528
|
NM_001130101
NM_005693
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr17_-_43394326
|
2.492
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr21_+_30671736
|
2.454
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr17_-_17184011
|
2.449
|
NM_001199125
|
COPS3
|
COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis)
|
|
chr12_+_104458235
|
2.419
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr15_-_34630203
|
2.418
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr17_-_48474910
|
2.410
|
NM_018509
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr2_+_208394615
|
2.409
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr13_+_20532788
|
2.401
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr19_+_11071792
|
2.349
|
NM_001128849
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr1_+_112938799
|
2.299
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr6_+_143929316
|
2.286
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr8_+_40010986
|
2.284
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr13_+_20532906
|
2.249
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr7_+_129015483
|
2.238
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_+_69644938
|
2.222
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr8_-_134072404
|
2.221
|
NM_006748
|
SLA
|
Src-like-adaptor
|
|
chr5_-_95297597
|
2.213
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr11_-_47400096
|
2.198
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr18_-_12884140
|
2.162
|
NM_001207013
NM_002828
NM_080422
NM_080423
|
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2
|
|
chr2_+_178077421
|
2.145
|
NM_194247
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr15_-_61521478
|
2.130
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_+_49832214
|
2.099
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr7_+_106809405
|
2.078
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr15_-_60919642
|
2.056
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_+_69644341
|
2.031
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr6_+_108881010
|
2.030
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr11_-_27721179
|
2.003
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_65430427
|
1.961
|
NM_001145138
NM_001243984
NM_001243985
NM_021975
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr16_-_20911560
|
1.950
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr5_+_112312392
|
1.949
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr6_+_118869441
|
1.941
|
NM_002667
|
PLN
|
phospholamban
|
|
chr17_-_40761404
|
1.926
|
NM_178126
|
FAM134C
|
family with sequence similarity 134, member C
|
|
chr1_+_2160133
|
1.924
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr17_-_56065596
|
1.914
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr22_+_40440664
|
1.891
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr2_-_128100673
|
1.879
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr20_+_51588945
|
1.872
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr6_+_163835668
|
1.863
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr8_-_71520620
|
1.846
|
NM_014294
|
TRAM1
|
translocation associated membrane protein 1
|
|
chr21_+_30671217
|
1.840
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr15_-_34629044
|
1.817
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_193091057
|
1.808
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr8_-_67525426
|
1.797
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr4_-_41216455
|
1.796
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr1_-_78444772
|
1.778
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr2_+_233925035
|
1.771
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chrX_+_107069081
|
1.757
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr9_-_16870719
|
1.738
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr14_+_96968698
|
1.724
|
NM_001252006
NM_001252007
NM_032632
|
PAPOLA
|
poly(A) polymerase alpha
|
|
chr14_+_58765209
|
1.698
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr10_+_86088156
|
1.692
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chrX_-_83442906
|
1.686
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr11_+_119076974
|
1.664
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr6_-_108145507
|
1.615
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr1_-_154842725
|
1.609
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_+_60280462
|
1.562
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr8_-_21771111
|
1.562
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr2_+_228336877
|
1.543
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr10_+_82213892
|
1.513
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr1_-_108507474
|
1.513
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_160283361
|
1.502
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr3_+_38495778
|
1.486
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr5_+_156607847
|
1.474
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr4_+_129730772
|
1.466
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr16_+_67063035
|
1.441
|
NM_001755
NM_022845
|
CBFB
|
core-binding factor, beta subunit
|
|
chr21_-_28217692
|
1.438
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr11_-_85780105
|
1.437
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_-_108231122
|
1.419
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr11_-_27721975
|
1.411
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr9_+_112542496
|
1.399
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr11_-_85780899
|
1.390
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_+_117824085
|
1.353
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr4_-_96470350
|
1.344
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr3_-_190040192
|
1.331
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr16_-_48644087
|
1.328
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr6_+_149639435
|
1.304
|
NM_015093
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr5_+_167718485
|
1.288
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr20_+_32150139
|
1.275
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr16_+_69599868
|
1.272
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr11_+_47270433
|
1.271
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr11_-_27681195
|
1.268
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr17_+_4613780
|
1.265
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chrX_+_129139163
|
1.258
|
NM_021946
|
BCORL1
|
BCL6 corepressor-like 1
|
|
chr2_-_39347562
|
1.257
|
NM_005633
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr13_+_112721912
|
1.239
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr6_+_108882068
|
1.226
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr5_-_148442606
|
1.217
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr11_-_27743604
|
1.209
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr16_-_51185150
|
1.165
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chrX_+_64708614
|
1.140
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr17_+_57642885
|
1.138
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr12_-_25055228
|
1.137
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr3_-_119812973
|
1.129
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr7_+_73624086
|
1.128
|
NM_014146
NM_032463
NM_032464
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr1_-_225840660
|
1.125
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr8_+_59465727
|
1.123
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr5_+_135468522
|
1.102
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr9_-_23826062
|
1.088
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr5_+_72112417
|
1.087
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr8_+_61591239
|
1.087
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr19_-_37019169
|
1.084
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr10_-_112064689
|
1.069
|
NM_005871
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr5_-_55290601
|
1.062
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr7_-_44924946
|
1.039
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr10_+_114709963
|
1.028
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr2_-_158731622
|
1.007
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr9_-_14314036
|
1.001
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr20_+_45523247
|
0.988
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr12_+_70133179
|
0.987
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr6_+_149068148
|
0.968
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr12_+_70132562
|
0.966
|
NM_022456
NM_175624
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|