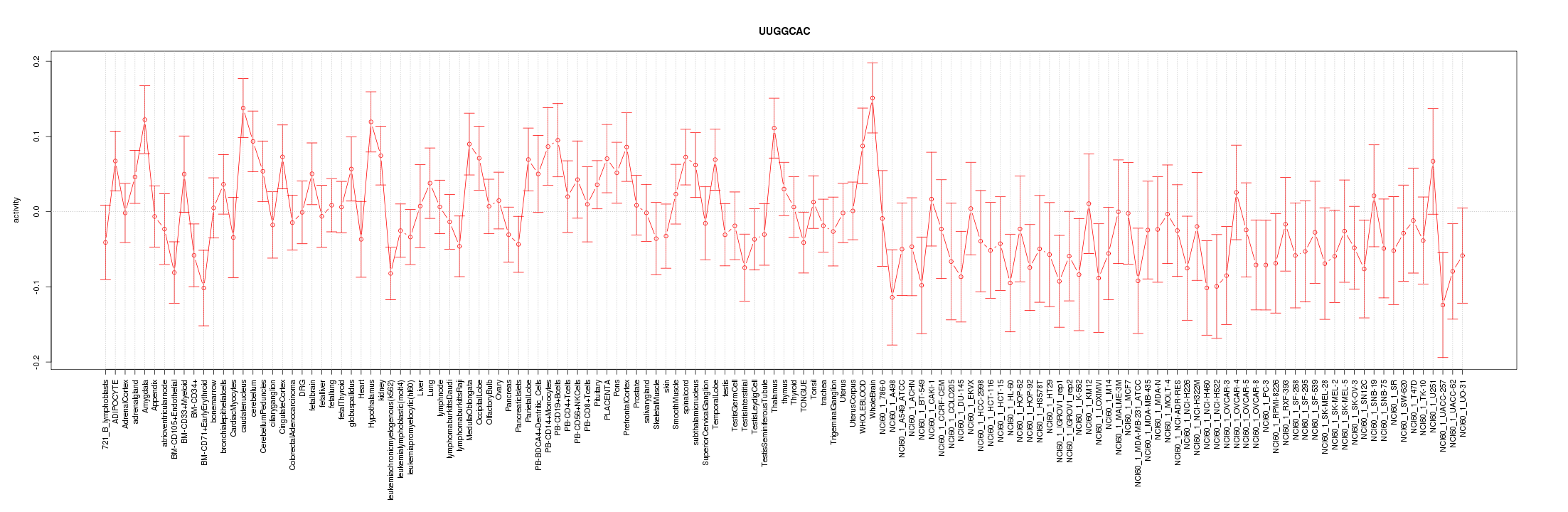
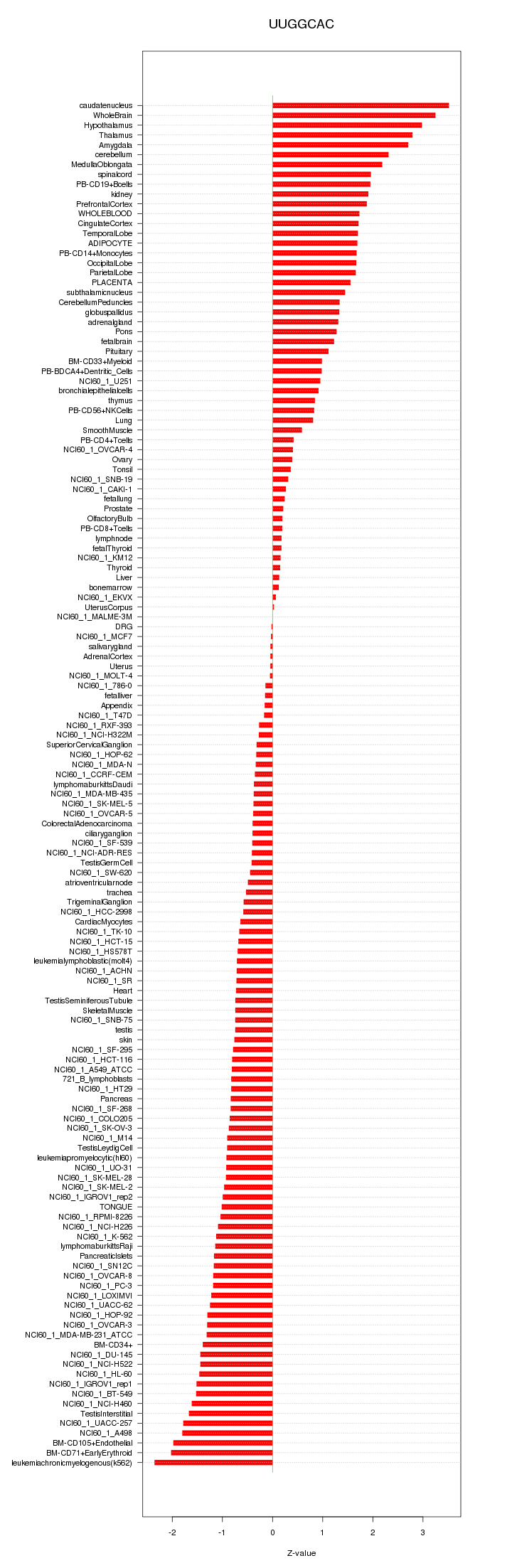
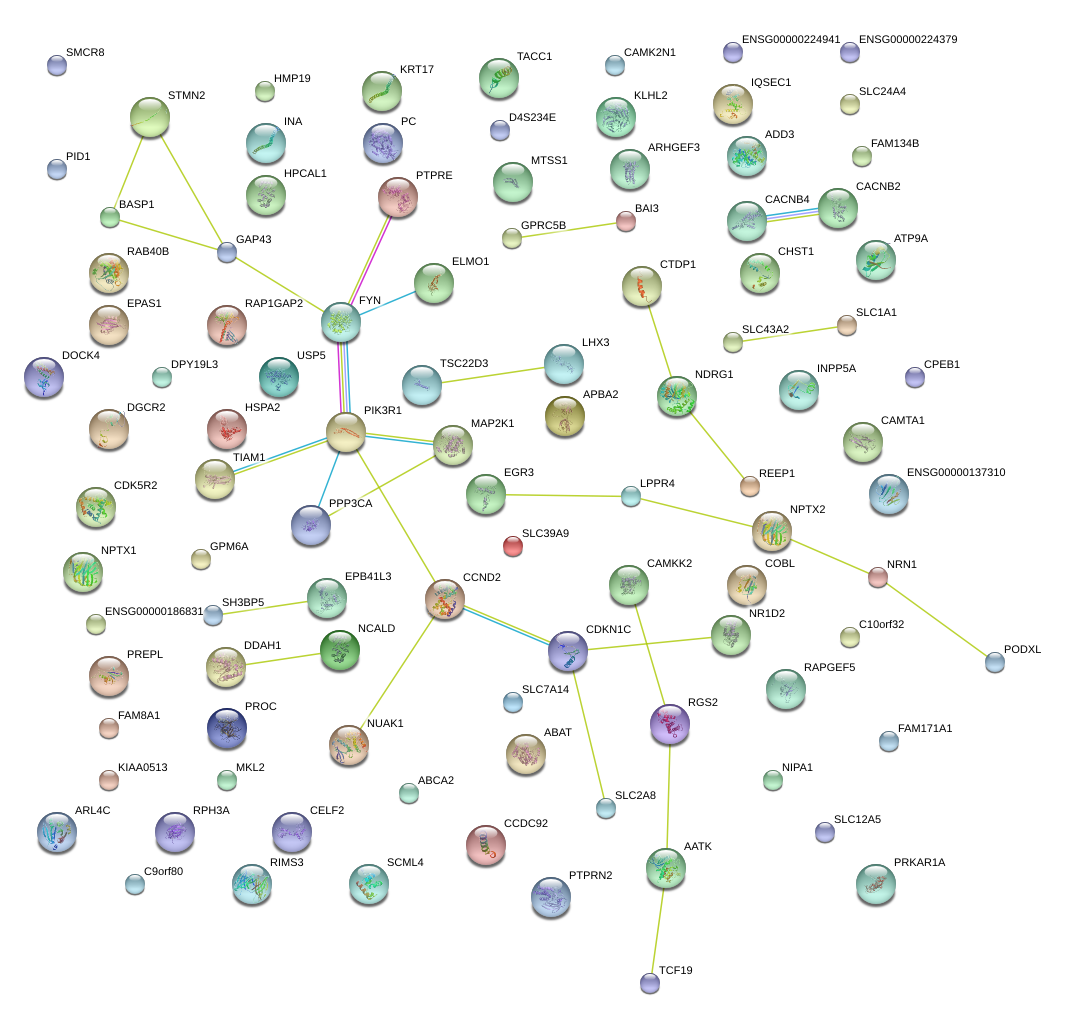
|
chr20_+_44650260
|
23.865
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr20_+_44657812
|
23.440
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr4_-_176734181
|
21.013
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr8_+_80523048
|
21.009
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr16_+_8768403
|
20.492
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr16_+_85061303
|
20.013
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr4_-_176923572
|
19.669
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr7_-_37393271
|
18.564
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr5_+_173472692
|
18.152
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr3_+_115342150
|
15.734
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr16_+_8806825
|
15.609
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr14_+_65007163
|
15.235
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chrX_-_107019001
|
14.985
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr7_-_37024664
|
14.902
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_-_79105747
|
14.807
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr16_+_8814567
|
14.570
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr3_-_13114547
|
14.351
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr7_-_37488554
|
14.134
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr12_-_106533810
|
14.127
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr7_-_111846384
|
13.996
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr15_-_23086290
|
13.810
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr7_-_37488894
|
13.614
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_-_79139871
|
13.554
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr6_-_6007632
|
13.453
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr20_-_50384899
|
13.292
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chrX_-_106960268
|
12.946
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_+_113229548
|
12.738
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr12_-_124457099
|
12.529
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr4_-_102268626
|
12.434
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr18_-_5543967
|
12.400
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr6_-_112080284
|
12.236
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr7_-_51384458
|
12.082
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr7_+_98246595
|
12.074
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr8_-_134309448
|
11.571
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr6_+_31126114
|
11.337
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|
|
chr3_-_15374082
|
11.055
|
NM_004844
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr6_-_112194648
|
10.951
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr11_-_66725790
|
10.935
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr6_-_112041253
|
10.856
|
NM_153047
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr3_-_15382874
|
10.718
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr11_-_45687135
|
10.706
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr10_+_105036783
|
10.622
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr15_+_29213839
|
10.569
|
NM_001130414
NM_005503
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2
|
|
chr5_+_17217474
|
10.562
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr9_-_139922705
|
10.526
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_-_20812727
|
10.487
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr2_-_44588943
|
9.985
|
NM_001171603
NM_001171617
|
PREPL
|
prolyl endopeptidase-like
|
|
chr8_-_103136486
|
9.850
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr9_-_139923214
|
9.766
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr17_-_80656461
|
9.753
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr8_-_125740650
|
9.702
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr1_-_85930733
|
9.597
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr14_+_92790151
|
9.493
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr8_-_103137134
|
9.444
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr16_-_19896105
|
9.415
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr17_-_78450247
|
9.312
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr10_-_15413054
|
9.211
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr7_-_22396511
|
9.153
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr15_-_83316621
|
9.010
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr2_-_235405692
|
8.965
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr16_+_14164879
|
8.948
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr10_+_129705299
|
8.929
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr7_-_158380360
|
8.868
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr4_+_4387982
|
8.829
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr8_-_102803438
|
8.800
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr3_-_57113292
|
8.735
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_+_23987514
|
8.698
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_-_44588622
|
8.677
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr9_+_4490193
|
8.490
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_-_44586854
|
8.481
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr10_+_129845806
|
8.385
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chrX_-_106959597
|
8.270
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_-_44587812
|
8.178
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr2_-_86565205
|
8.174
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr17_+_2699731
|
8.130
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr1_+_99729847
|
8.117
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr6_-_108145507
|
8.078
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr10_+_134351272
|
7.987
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr5_+_67584251
|
7.938
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_-_2906960
|
7.930
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr1_+_192778168
|
7.891
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr2_-_86564632
|
7.868
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr6_+_17600504
|
7.815
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr4_+_166128769
|
7.801
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr22_-_19109912
|
7.646
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr17_+_18218569
|
7.588
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr5_-_16617085
|
7.582
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_+_32896515
|
7.427
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr10_+_11059854
|
7.407
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr17_+_66508098
|
7.406
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr18_+_77439798
|
7.401
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr10_+_111767685
|
7.398
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr14_+_69865380
|
7.395
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr21_-_32931289
|
7.316
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr9_-_115480363
|
7.302
|
NM_021218
|
C9orf80
|
chromosome 9 open reading frame 80
|
|
chr14_+_92789536
|
7.212
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr2_+_219824346
|
7.166
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr2_-_152830448
|
7.024
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr19_-_33793429
|
6.997
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr10_+_104613966
|
6.976
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr8_+_38644721
|
6.947
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr11_-_66675334
|
6.606
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr12_+_4382882
|
6.595
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr17_-_1532109
|
6.502
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr5_-_16509106
|
6.482
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr9_-_139094886
|
6.466
|
NM_014564
|
LHX3
|
LIM homeobox 3
|
|
chr15_+_66679107
|
6.462
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr1_+_6845381
|
6.458
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr4_+_166131170
|
6.450
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr12_+_6961257
|
6.339
|
NM_001098536
NM_003481
|
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T)
|
|
chr3_-_170303774
|
6.248
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr5_+_67511578
|
6.197
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr10_+_18429605
|
6.187
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr2_-_230135954
|
6.170
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr3_-_56835829
|
6.167
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_-_22550708
|
6.136
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_-_41131323
|
6.106
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr2_+_46524448
|
6.100
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr12_-_121734543
|
6.053
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr6_+_69344939
|
6.052
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr9_+_130159383
|
6.045
|
NM_014580
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8
|
|
chr13_+_22245083
|
6.036
|
NM_002010
|
FGF9
|
fibroblast growth factor 9 (glia-activating factor)
|
|
chr8_+_61429390
|
6.003
|
NM_001242644
NM_002865
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr10_+_11047253
|
5.987
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr13_-_41240607
|
5.987
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr1_+_205473683
|
5.927
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr10_-_75910525
|
5.926
|
NM_012095
NM_207012
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr7_+_87257704
|
5.903
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr6_+_138483024
|
5.898
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr14_-_53620032
|
5.894
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr10_+_95517565
|
5.889
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chrX_-_13835313
|
5.871
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr8_+_38585703
|
5.769
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr18_+_11851388
|
5.749
|
NM_020412
|
CHMP1B
|
charged multivesicular body protein 1B
|
|
chr16_-_29910340
|
5.737
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr19_-_10341947
|
5.708
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr5_+_76506705
|
5.694
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr3_-_56502364
|
5.684
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr8_-_22549901
|
5.669
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr5_-_115910406
|
5.668
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr22_-_39268230
|
5.644
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr16_-_29937539
|
5.640
|
NM_178863
|
KCTD13
|
potassium channel tetramerisation domain containing 13
|
|
chr6_-_91006460
|
5.622
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_+_51632507
|
5.598
|
NM_001136264
NM_001136266
NM_001136267
NM_001136268
NM_001136269
NM_014764
|
DAZAP2
|
DAZ associated protein 2
|
|
chr12_-_42538432
|
5.591
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr3_-_9994057
|
5.571
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr1_-_86043932
|
5.498
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_+_138066489
|
5.492
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr17_+_73043154
|
5.491
|
NM_015353
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr5_-_171433876
|
5.490
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr12_+_77157842
|
5.391
|
NM_015336
|
ZDHHC17
|
zinc finger, DHHC-type containing 17
|
|
chr8_+_58906968
|
5.344
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr12_-_121736110
|
5.329
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr12_+_121078421
|
5.320
|
NM_001033677
|
CABP1
|
calcium binding protein 1
|
|
chr4_-_42658883
|
5.294
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr17_+_66508525
|
5.224
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr1_-_155532322
|
5.202
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr6_-_91006580
|
5.178
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr15_-_52861177
|
5.165
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr13_+_39612440
|
5.131
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr13_-_53422773
|
5.124
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr10_+_104678001
|
5.122
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr10_+_18549583
|
5.108
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr4_-_88141614
|
5.101
|
NM_020803
|
KLHL8
|
kelch-like 8 (Drosophila)
|
|
chr5_-_58334936
|
5.085
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_152955208
|
5.070
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_+_67588395
|
5.032
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_31799030
|
5.016
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr17_+_68165660
|
5.002
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_-_101034049
|
4.982
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr3_-_56809594
|
4.981
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_110527327
|
4.978
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr5_+_67586466
|
4.975
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_+_30594607
|
4.941
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr12_+_121088370
|
4.938
|
NM_004276
NM_031205
|
CABP1
|
calcium binding protein 1
|
|
chr1_-_177133950
|
4.905
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr6_-_6320897
|
4.879
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr19_+_16940172
|
4.878
|
NM_015260
|
SIN3B
|
SIN3 transcription regulator homolog B (yeast)
|
|
chr7_+_97361270
|
4.821
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr1_-_109584589
|
4.787
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr11_-_115375065
|
4.779
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr2_-_166930130
|
4.745
|
NM_001165963
NM_001165964
NM_006920
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr1_+_110543548
|
4.722
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr14_+_92788924
|
4.711
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr19_-_6279927
|
4.711
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr3_-_66550844
|
4.699
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chrX_-_102941582
|
4.666
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr6_-_169654047
|
4.665
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr18_+_56530060
|
4.610
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr2_+_203499819
|
4.604
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr9_+_19049355
|
4.589
|
NM_006570
|
RRAGA
|
Ras-related GTP binding A
|
|
chrX_+_70315637
|
4.588
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chrX_+_14903412
|
4.575
|
NM_001177475
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr3_-_57678594
|
4.572
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr16_+_50776012
|
4.567
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr12_-_56615679
|
4.565
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr5_-_179498995
|
4.559
|
NM_018434
|
RNF130
|
ring finger protein 130
|
|
chr2_+_113033146
|
4.527
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr2_-_197036301
|
4.509
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr2_-_96931744
|
4.502
|
NM_001193304
NM_017849
|
TMEM127
|
transmembrane protein 127
|