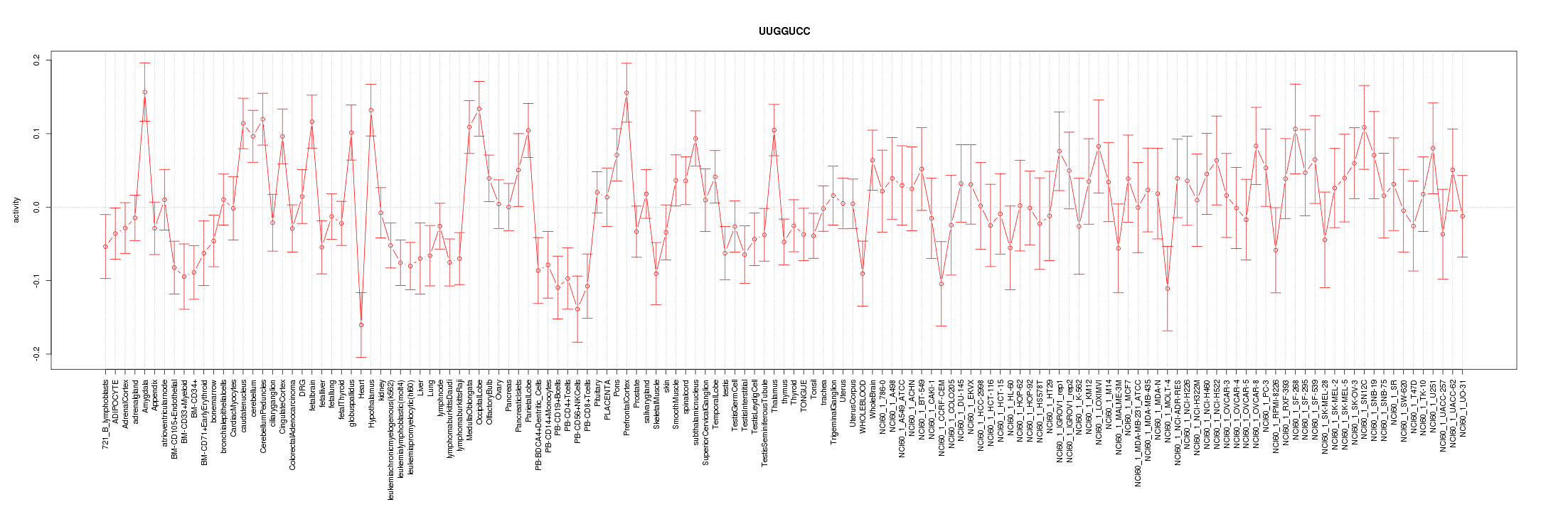
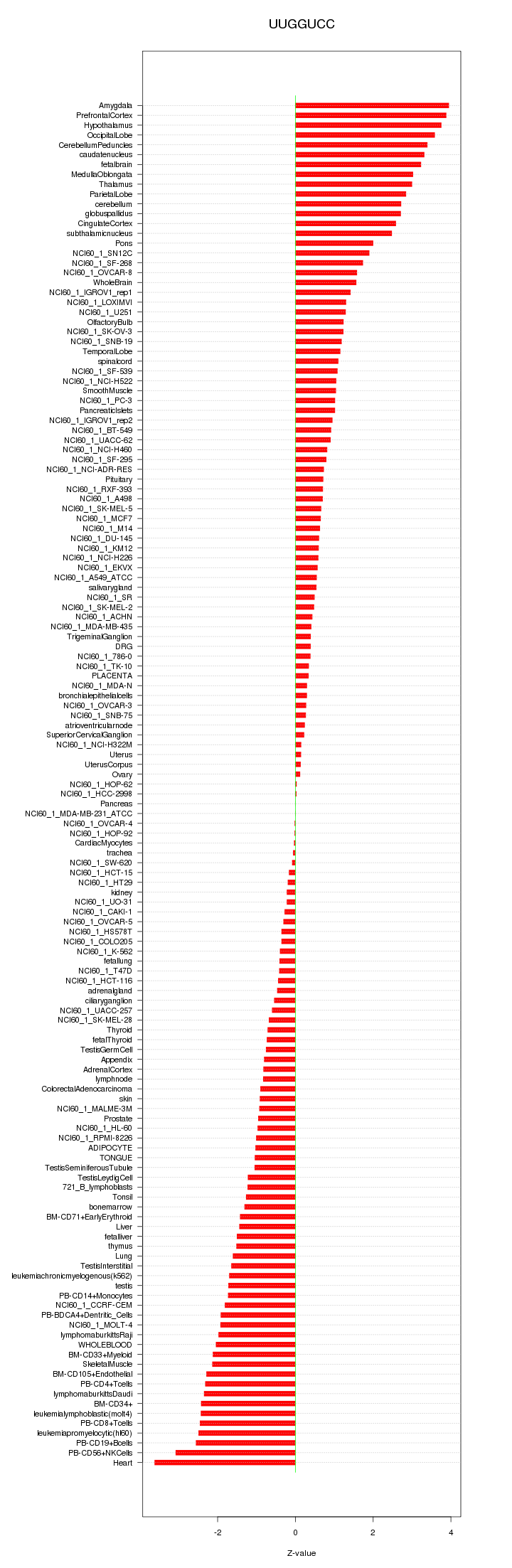
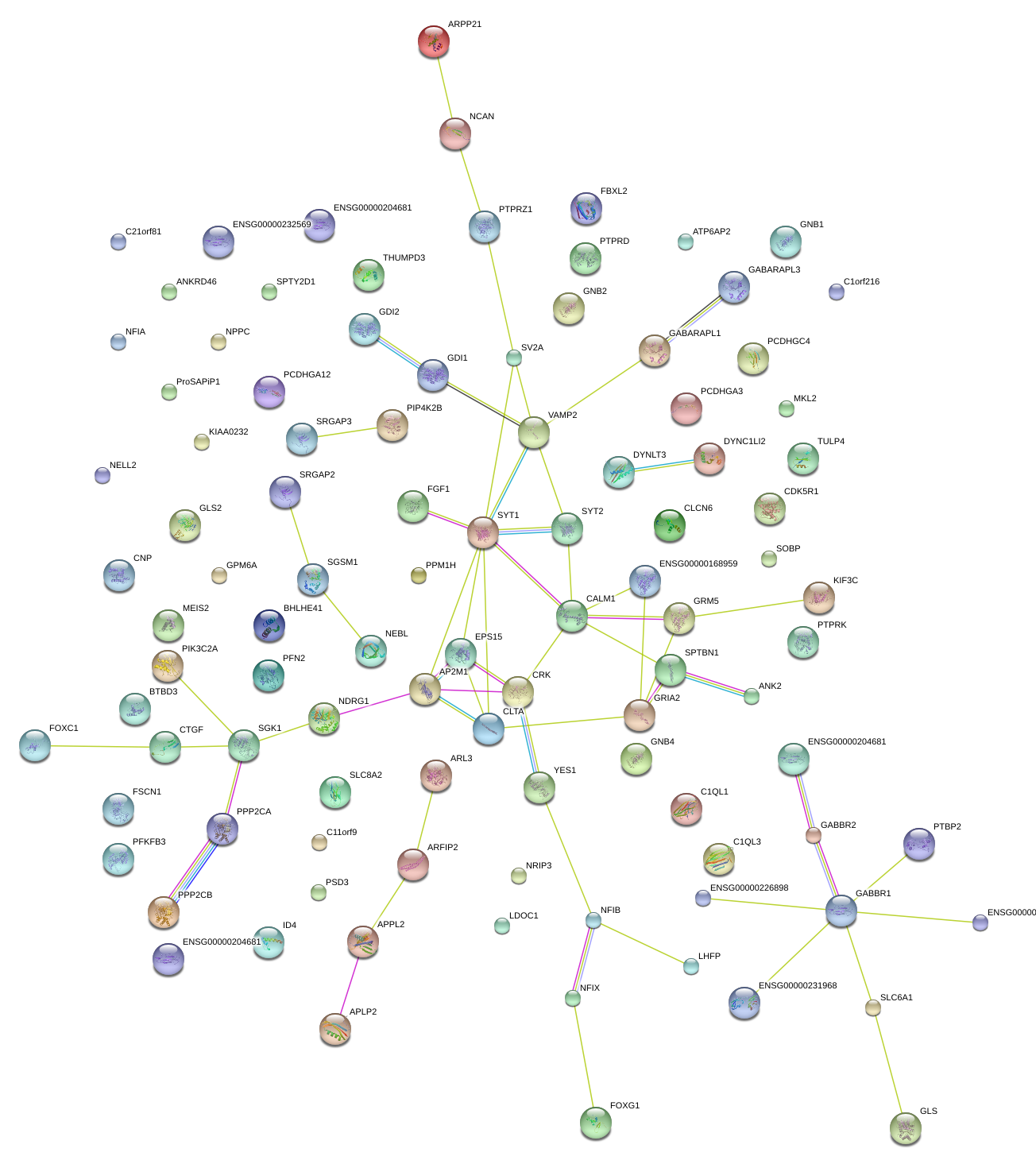
|
chr3_-_149688638
|
31.862
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr4_-_176923572
|
30.273
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_176734181
|
27.648
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr12_+_79439432
|
22.868
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr20_+_11871364
|
22.795
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr12_+_79257772
|
22.771
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr7_+_121513158
|
21.766
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr12_+_79258448
|
21.557
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr6_+_107811272
|
21.524
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr1_-_149889362
|
21.024
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr8_-_18871163
|
20.482
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_+_113739203
|
20.417
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr17_+_40118696
|
17.307
|
NM_033133
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chrX_-_140271293
|
17.091
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr20_+_11898536
|
17.014
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr4_+_158141735
|
16.307
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr16_-_66785511
|
15.561
|
NM_006141
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chr5_-_142065586
|
15.347
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_-_18666295
|
15.181
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chrX_+_153665258
|
15.162
|
NM_001493
|
GDI1
|
GDP dissociation inhibitor 1
|
|
chr5_-_142065991
|
15.014
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr9_-_101470836
|
14.570
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr4_+_113970784
|
14.298
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr22_+_25202135
|
13.804
|
NM_001039948
NM_001098497
NM_001098498
NM_133454
|
SGSM1
|
small G protein signaling modulator 1
|
|
chr5_-_142077508
|
13.735
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_-_26205437
|
13.517
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr3_+_11034419
|
13.497
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr14_+_90863302
|
13.272
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr16_+_14164879
|
13.075
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr20_-_3149070
|
12.992
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr12_+_10365415
|
12.901
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr13_-_40177355
|
12.052
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr5_-_142000899
|
11.725
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_-_134309448
|
11.175
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr15_-_37393364
|
10.882
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr5_-_141993691
|
10.828
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_-_128841432
|
10.682
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr11_+_61522860
|
10.435
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr19_-_47975244
|
10.260
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr9_-_10612722
|
9.653
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr12_-_26277807
|
9.390
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr12_-_45269340
|
9.156
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr12_-_45307710
|
9.044
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr11_-_9025531
|
8.862
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr3_+_183892569
|
8.854
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr12_-_45270632
|
8.820
|
NM_001145107
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr15_-_37392358
|
8.765
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr5_-_133561805
|
8.759
|
NM_002715
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr12_-_45270157
|
8.578
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr19_+_19322763
|
8.513
|
NM_004386
|
NCAN
|
neurocan
|
|
chr11_+_61520000
|
8.394
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr6_+_158733691
|
8.352
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr15_-_37390372
|
8.140
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr17_-_43045612
|
7.765
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr7_+_5632435
|
7.613
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr2_+_191745546
|
7.491
|
NM_014905
|
GLS
|
glutaminase
|
|
chr3_-_9291251
|
7.382
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr6_+_19837599
|
7.154
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr9_-_8733893
|
7.121
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr1_-_36184727
|
6.992
|
NM_152374
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr6_-_134498973
|
6.965
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_-_104473957
|
6.832
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr6_-_134496006
|
6.786
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_+_30813928
|
6.747
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr11_-_6502563
|
6.710
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr8_-_30670323
|
6.617
|
NM_001009552
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr1_+_97187174
|
6.605
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr5_+_140739589
|
6.550
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr5_+_140749830
|
6.485
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr8_-_101571870
|
6.458
|
NM_198401
|
GAPDHP62
ANKRD46
|
glyceraldehyde 3 phosphate dehydrogenase pseudogene 62
ankyrin repeat domain 46
|
|
chr4_+_6784453
|
6.446
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr18_-_812268
|
6.392
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr9_+_36190852
|
6.311
|
NM_001076677
NM_001184760
NM_001184761
NM_001184762
NM_001833
NM_007096
|
CLTA
|
clathrin, light chain A
|
|
chr6_-_132272311
|
6.282
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr5_+_140864626
|
6.281
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr12_-_63328663
|
6.255
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr9_-_14398981
|
6.186
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_202612580
|
6.144
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr3_+_33318911
|
6.132
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr11_-_18655962
|
6.092
|
NM_194285
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr5_+_140868721
|
6.031
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr3_+_9404706
|
6.022
|
NM_001114092
NM_015453
|
THUMPD3
|
THUMP domain containing 3
|
|
chr10_-_21463019
|
5.954
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr2_+_54683276
|
5.924
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr5_+_140797243
|
5.832
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr15_-_37391596
|
5.828
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr3_+_35722428
|
5.709
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr11_-_17191287
|
5.697
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr5_+_140729723
|
5.668
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr6_-_29595934
|
5.666
|
NM_021903
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr14_+_29236171
|
5.664
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr17_-_36956145
|
5.659
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr6_+_1610680
|
5.644
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chr6_-_29600959
|
5.595
|
NM_001470
NM_021904
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr17_-_8066254
|
5.523
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr17_-_1359410
|
5.497
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chrX_-_37706816
|
5.464
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr6_-_134497069
|
5.355
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_+_6186758
|
5.293
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_-_51984994
|
5.198
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr11_-_88781039
|
5.174
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chrX_+_40440145
|
5.108
|
NM_005765
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr12_-_105629790
|
5.096
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr3_-_179169330
|
5.093
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr1_+_11866206
|
5.088
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr4_+_26321307
|
5.081
|
NM_005349
NM_203283
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr6_-_143266283
|
5.035
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr8_-_103137134
|
4.923
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr9_-_139923214
|
4.892
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr7_-_37393271
|
4.770
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr8_+_22423170
|
4.762
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr3_-_15839674
|
4.752
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chrX_-_48900932
|
4.690
|
NM_006521
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr9_-_14314036
|
4.663
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr3_+_35721115
|
4.537
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr1_+_165796652
|
4.533
|
NM_012474
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr15_+_85525204
|
4.530
|
NM_002605
NM_173454
|
PDE8A
|
phosphodiesterase 8A
|
|
chr1_+_151043079
|
4.504
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr6_+_125475334
|
4.484
|
NM_001003395
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr7_-_37024664
|
4.417
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_-_102605974
|
4.410
|
NM_001017963
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1
|
|
chr10_-_98346793
|
4.382
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_-_134639195
|
4.372
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_-_171433876
|
4.342
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chrX_+_16737661
|
4.325
|
NM_032796
|
SYAP1
|
synapse associated protein 1
|
|
chr11_-_117166247
|
4.263
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr9_-_21335239
|
4.260
|
NM_018847
|
KLHL9
|
kelch-like 9 (Drosophila)
|
|
chr13_-_33760215
|
4.214
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_118047336
|
4.201
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr8_-_102803438
|
4.141
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr16_+_5008317
|
4.128
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr9_+_130374466
|
4.104
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr4_-_39640461
|
4.101
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr8_-_103136486
|
4.090
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr9_-_139922705
|
4.084
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr11_-_88796815
|
4.071
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr8_-_38326351
|
4.056
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr7_-_22396511
|
4.033
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr14_-_102553386
|
3.972
|
NM_005348
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1
|
|
chr6_+_125474800
|
3.943
|
NM_001003396
NM_001003397
NM_003287
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr17_-_40307015
|
3.855
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr11_-_61584451
|
3.851
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr5_+_118407067
|
3.840
|
NM_005509
|
DMXL1
|
Dmx-like 1
|
|
chr7_-_82792127
|
3.823
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr17_+_5031686
|
3.820
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr12_-_42538432
|
3.819
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr5_+_140810126
|
3.818
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr5_+_140753480
|
3.814
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr12_+_110719029
|
3.814
|
NM_001681
NM_170665
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr4_-_7941092
|
3.775
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr13_-_33859835
|
3.774
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_7831326
|
3.754
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr7_-_158622202
|
3.744
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr4_+_166128769
|
3.720
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr11_+_35639734
|
3.717
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr14_+_23775970
|
3.716
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr7_-_37488894
|
3.696
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_-_14316980
|
3.696
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr10_-_21186530
|
3.681
|
NM_006393
|
NEBL
|
nebulette
|
|
chr9_+_34958171
|
3.660
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr3_-_133614421
|
3.643
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr15_+_41851153
|
3.587
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr15_-_23086290
|
3.574
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr14_+_69658482
|
3.551
|
NM_001193362
NM_001193363
|
EXD2
|
exonuclease 3'-5' domain containing 2
|
|
chr14_+_69658193
|
3.524
|
NM_001193360
NM_001193361
NM_018199
|
EXD2
|
exonuclease 3'-5' domain containing 2
|
|
chr8_-_53626991
|
3.511
|
NM_001083617
NM_014781
|
RB1CC1
|
RB1-inducible coiled-coil 1
|
|
chr3_-_15900928
|
3.491
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr18_-_78005230
|
3.473
|
NM_032510
|
PARD6G
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr3_-_15798057
|
3.455
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_117186918
|
3.420
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr4_+_86396251
|
3.414
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr6_-_80656870
|
3.399
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr5_+_140743755
|
3.309
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr4_+_26322184
|
3.252
|
NM_015874
NM_203284
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr2_+_177053306
|
3.241
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr4_+_166131170
|
3.238
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr3_-_45267804
|
3.180
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr9_-_138987096
|
3.153
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr3_-_160822625
|
3.041
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr14_+_103243813
|
3.038
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr5_+_175875355
|
3.032
|
NM_014613
|
FAF2
|
Fas associated factor family member 2
|
|
chr8_+_22409226
|
2.998
|
NM_005775
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr9_+_131873592
|
2.996
|
NM_178000
NM_178001
NM_178003
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr17_+_53342320
|
2.981
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr5_+_140767402
|
2.977
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr14_+_105331581
|
2.961
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr1_-_202679415
|
2.958
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr2_-_73298815
|
2.958
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr12_-_82153092
|
2.939
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_+_78245306
|
2.917
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr1_+_201924618
|
2.856
|
NM_006335
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast)
|
|
chr8_-_66701175
|
2.850
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr22_-_28315214
|
2.840
|
NM_012399
|
PITPNB
|
phosphatidylinositol transfer protein, beta
|
|
chr17_+_635656
|
2.828
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chr17_+_73043154
|
2.795
|
NM_015353
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr12_-_121736110
|
2.787
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr14_-_25519076
|
2.765
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr12_-_109458844
|
2.753
|
NM_018711
|
SVOP
|
SV2 related protein homolog (rat)
|
|
chr10_+_71211215
|
2.748
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr8_+_38034066
|
2.733
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|