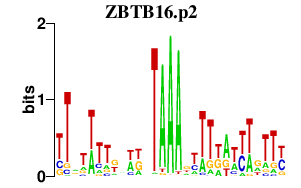
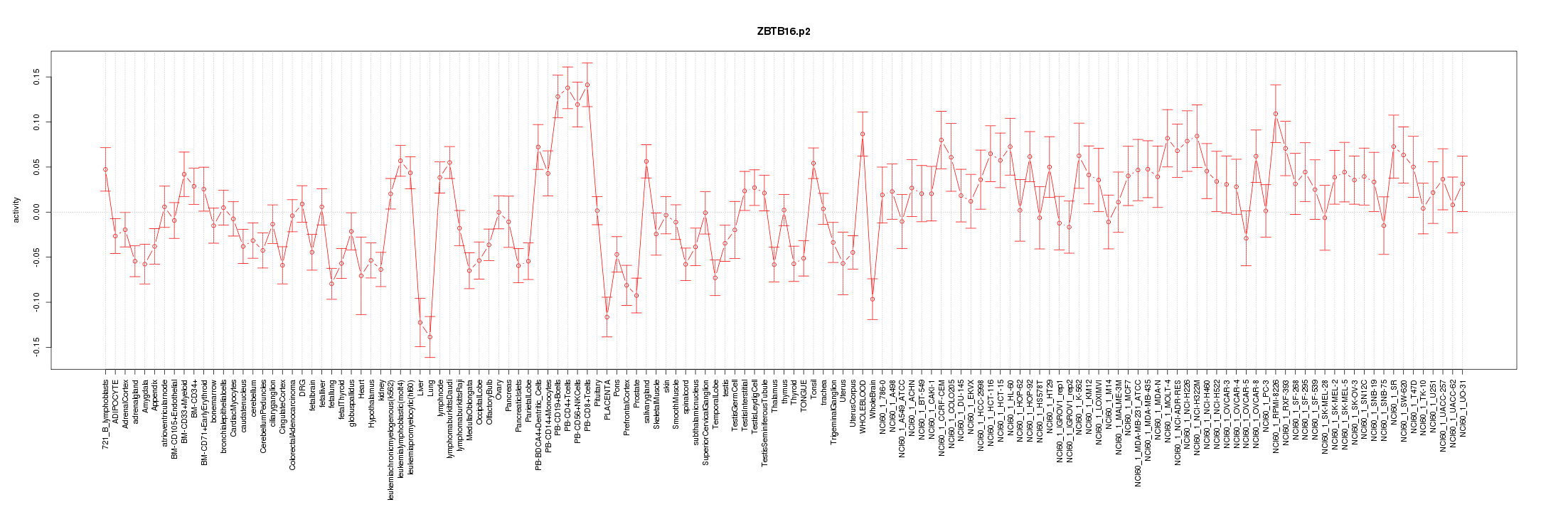
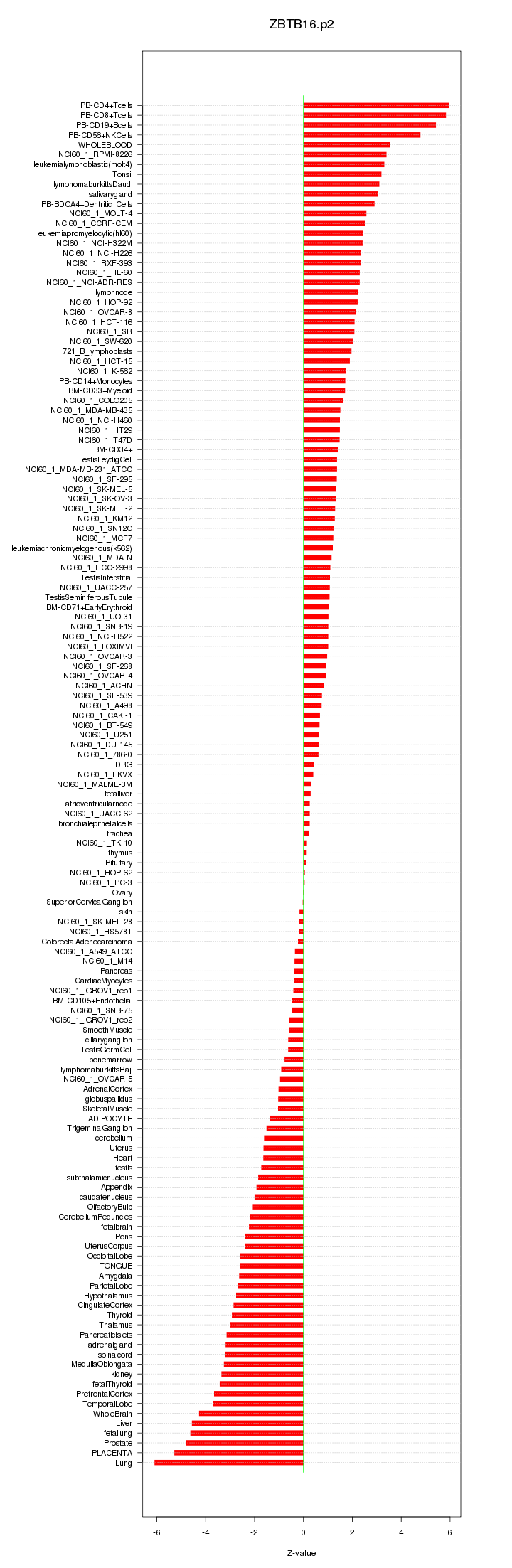
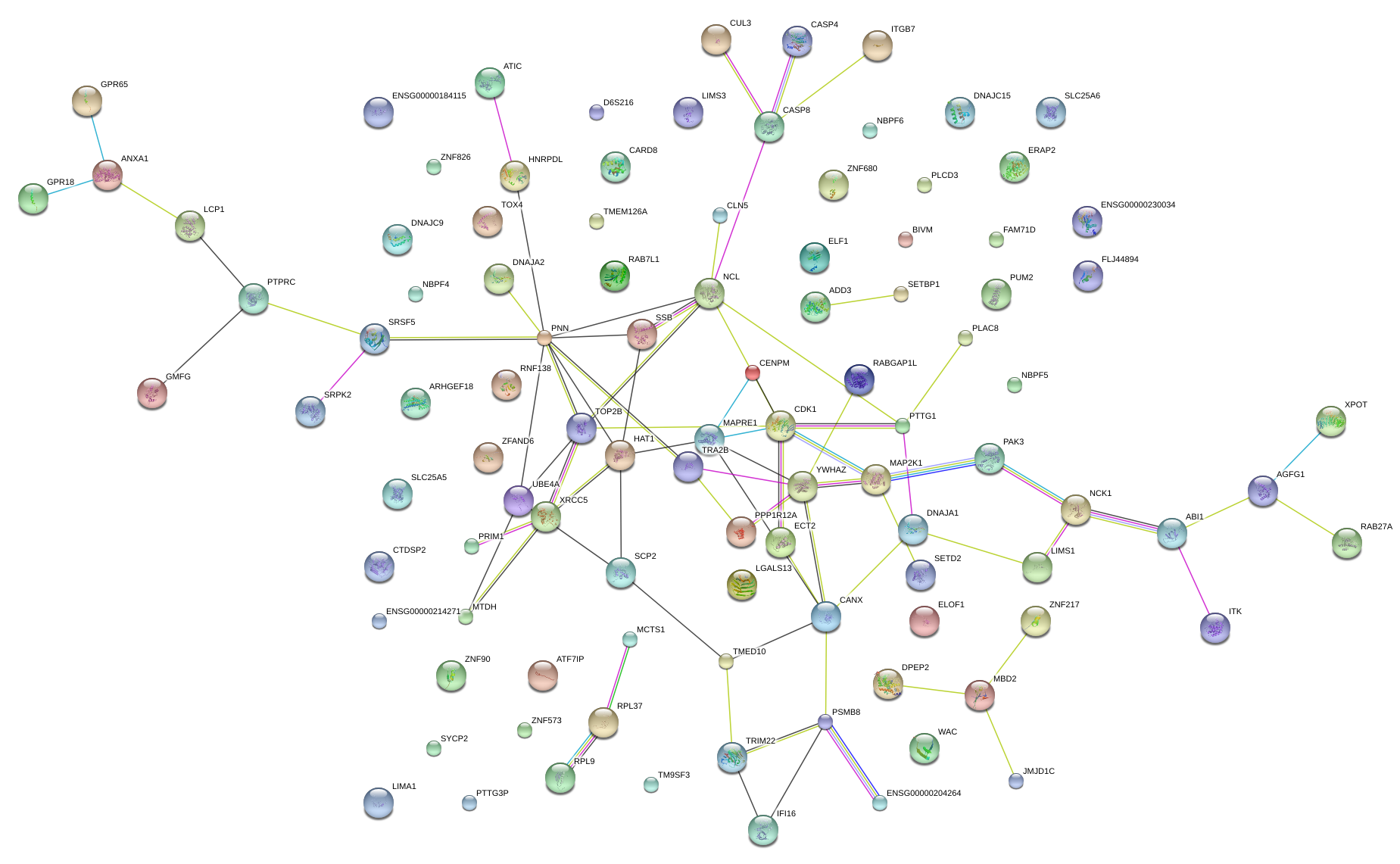
|
chr1_+_198608136
|
45.532
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr19_-_20607761
|
44.320
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr14_+_39649711
|
42.700
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr12_-_53601051
|
41.220
|
|
ITGB7
|
integrin, beta 7
|
|
chr1_+_198608216
|
38.867
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_108786663
|
38.699
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chr10_-_98325100
|
38.594
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_+_198608244
|
37.679
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_-_225370687
|
34.041
|
|
CUL3
|
cullin 3
|
|
chr1_+_174844655
|
33.387
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr13_-_99910548
|
30.548
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr10_+_62538088
|
30.254
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr3_-_185641596
|
29.998
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr11_-_67450536
|
29.931
|
|
RPL37
|
ribosomal protein L37
|
|
chr19_-_48752921
|
29.767
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr10_+_111765725
|
28.466
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr3_+_136649316
|
28.224
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr19_+_20277991
|
27.790
|
NM_052852
|
ZNF486
|
zinc finger protein 486
|
|
chr11_-_104827421
|
26.637
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr18_-_51690988
|
26.442
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr17_+_26794838
|
25.691
|
|
|
|
|
chr12_-_80307690
|
25.689
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr3_+_172472246
|
25.666
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr10_-_27057776
|
25.587
|
|
ABI1
|
abl-interactor 1
|
|
chr5_+_96212167
|
25.431
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr2_+_172821874
|
24.736
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr19_-_48759141
|
24.580
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr12_-_53600999
|
24.504
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr11_+_5710816
|
24.187
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr14_+_67656109
|
23.550
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr14_+_70236266
|
23.343
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr2_-_20512016
|
23.197
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr1_+_53480573
|
22.937
|
NM_001007099
NM_001007100
NM_001007250
|
SCP2
|
sterol carrier protein 2
|
|
chr5_+_159848876
|
22.747
|
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr10_-_65028825
|
22.658
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr5_+_179132676
|
22.474
|
|
CANX
|
calnexin
|
|
chr14_-_75600111
|
22.293
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr5_+_156607847
|
22.068
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr2_+_170662044
|
21.680
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr5_+_159848835
|
21.140
|
NM_004219
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr12_+_14538232
|
20.689
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr2_+_109237679
|
19.855
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr4_-_84035888
|
19.699
|
|
PLAC8
|
placenta-specific 8
|
|
chr4_-_84035908
|
19.548
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr13_-_41556360
|
18.572
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr13_+_43597338
|
17.936
|
NM_013238
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15
|
|
chrX_-_23855456
|
17.402
|
|
RPL9
|
ribosomal protein L9
|
|
chr15_-_55562483
|
17.353
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr7_-_64023331
|
17.223
|
|
ZNF680
|
zinc finger protein 680
|
|
chrX_+_119737743
|
17.177
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr20_-_52199635
|
17.164
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr12_-_46357949
|
16.966
|
|
|
|
|
chr15_+_80390757
|
16.861
|
NM_001242919
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr15_+_80364902
|
16.711
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr8_-_101936521
|
16.598
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr2_+_202125222
|
16.296
|
NM_033356
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr12_-_57146081
|
16.286
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr11_+_118257222
|
16.118
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr3_-_47098906
|
16.037
|
|
SETD2
|
SET domain containing 2
|
|
chr13_-_41593405
|
15.830
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr19_-_21646685
|
15.820
|
|
|
|
|
chr1_+_158979681
|
15.761
|
NM_001206567
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr16_-_47005339
|
15.731
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr9_+_75766777
|
15.654
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr7_-_64023454
|
15.474
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr1_-_205744543
|
15.089
|
NM_001135662
NM_001135663
NM_001135664
NM_003929
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr18_+_29671817
|
15.084
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr10_+_28878666
|
15.056
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr2_+_228356287
|
15.050
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr1_+_53480616
|
15.031
|
|
SCP2
|
sterol carrier protein 2
|
|
chr8_+_98703361
|
14.900
|
|
MTDH
|
metadherin
|
|
chrX_+_118602362
|
14.587
|
NM_001152
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr4_-_83347778
|
14.484
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr2_-_232326394
|
14.456
|
|
NCL
|
nucleolin
|
|
chr3_-_25683825
|
14.398
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr15_+_66694216
|
14.334
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr7_-_104909442
|
14.144
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chrX_+_118602391
|
14.141
|
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr12_+_64803741
|
14.077
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr19_+_7504573
|
13.959
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr13_-_46756291
|
13.871
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr10_-_75006963
|
13.853
|
NM_015190
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9
|
|
chr13_+_103459495
|
13.800
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr2_+_216176678
|
13.728
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr2_+_216974101
|
13.597
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chr20_+_31407696
|
13.577
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr19_-_39826676
|
13.506
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr14_+_88471467
|
13.487
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr22_-_42336222
|
13.444
|
NM_001110215
|
CENPM
|
centromere protein M
|
|
chr6_-_32812711
|
13.259
|
NM_004159
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr9_+_75766794
|
13.252
|
|
ANXA1
|
annexin A1
|
|
chr12_-_50616425
|
13.194
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr2_-_150444219
|
12.931
|
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr1_+_43162386
|
12.844
|
|
YBX1
|
Y box binding protein 1
|
|
chr13_-_41593491
|
12.679
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr9_+_79968344
|
12.635
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr2_+_216974006
|
12.589
|
NM_021141
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chr11_+_71249070
|
12.522
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr4_-_83347222
|
12.422
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr1_+_100731797
|
12.365
|
|
RTCD1
|
RNA terminal phosphate cyclase domain 1
|
|
chr14_+_58906398
|
12.303
|
NM_001244193
|
KIAA0586
|
KIAA0586
|
|
chr14_-_20797532
|
12.216
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr7_+_142960502
|
12.162
|
NM_001143679
NM_001143680
NM_001143681
NM_015917
|
GSTK1
|
glutathione S-transferase kappa 1
|
|
chr12_+_10460416
|
12.130
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr3_-_146262421
|
12.107
|
NM_021105
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr10_+_51837885
|
12.102
|
NM_018232
|
FAM21B
|
family with sequence similarity 21, member B
|
|
chr13_-_37573474
|
11.951
|
NM_001142364
NM_013338
|
ALG5
|
asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae)
|
|
chrX_+_119737905
|
11.901
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr8_-_80993009
|
11.889
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr12_+_70636773
|
11.881
|
NM_014515
NM_001199302
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr2_-_150444236
|
11.876
|
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr15_-_83018159
|
11.729
|
NM_001164465
|
LOC643696
GOLGA6L10
|
uncharacterized LOC643696
golgin A6 family-like 10
|
|
chr2_+_161993465
|
11.692
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr7_+_142960588
|
11.645
|
|
GSTK1
|
glutathione S-transferase kappa 1
|
|
chr2_-_100046262
|
11.611
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr13_-_37573435
|
11.539
|
|
ALG5
|
asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae)
|
|
chr1_-_108735362
|
11.401
|
NM_213651
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24
|
|
chr2_-_47134945
|
11.341
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr1_-_147624266
|
11.296
|
|
NBPF11
|
neuroblastoma breakpoint family, member 11
|
|
chr2_-_74382352
|
11.288
|
|
|
|
|
chr11_+_5710926
|
11.133
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr19_+_13134782
|
10.926
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr5_+_68667280
|
10.894
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr2_+_183608329
|
10.883
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr17_-_3599298
|
10.875
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr16_-_85722503
|
10.658
|
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr12_+_69742130
|
10.607
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr2_-_150444285
|
10.533
|
NM_015702
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr20_+_52824616
|
10.474
|
|
PFDN4
|
prefoldin subunit 4
|
|
chr5_+_132428394
|
10.386
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr6_-_99849266
|
10.380
|
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr1_-_111743297
|
10.296
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr10_+_47894022
|
10.246
|
NM_018232
|
FAM21B
|
family with sequence similarity 21, member B
|
|
chr15_-_82641705
|
10.226
|
NM_001164465
|
GOLGA6L10
|
golgin A6 family-like 10
|
|
chr5_-_98262217
|
10.025
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr4_+_1953831
|
10.002
|
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr4_+_15779887
|
9.951
|
NM_001775
|
CD38
|
CD38 molecule
|
|
chr14_+_21457964
|
9.929
|
NM_001029991
NM_022734
|
METTL17
|
methyltransferase like 17
|
|
chr12_+_118454505
|
9.834
|
NM_001130112
NM_001206801
NM_007370
NM_181578
|
RFC5
|
replication factor C (activator 1) 5, 36.5kDa
|
|
chr17_+_46908371
|
9.832
|
NM_005831
|
CALCOCO2
|
calcium binding and coiled-coil domain 2
|
|
chr16_-_85722587
|
9.739
|
NM_016095
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr11_-_47510518
|
9.720
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr1_+_100731696
|
9.716
|
NM_001130841
NM_003729
|
RTCD1
|
RNA terminal phosphate cyclase domain 1
|
|
chr2_+_234296799
|
9.632
|
NM_003648
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr12_+_54892567
|
9.627
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr5_+_35856990
|
9.572
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr10_+_70847817
|
9.563
|
NM_002727
|
SRGN
|
serglycin
|
|
chr13_-_41556188
|
9.499
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr2_+_170655321
|
9.489
|
NM_003142
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr8_+_74903563
|
9.467
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr10_+_5772610
|
9.412
|
|
FAM208B
|
family with sequence similarity 208, member B
|
|
chr11_-_1629614
|
9.287
|
NM_001012708
|
KRTAP5-3
|
keratin associated protein 5-3
|
|
chr6_-_100016517
|
9.287
|
NM_001013399
NM_005190
|
CCNC
|
cyclin C
|
|
chr12_+_32832128
|
9.181
|
NM_005690
NM_012062
NM_012063
|
DNM1L
|
dynamin 1-like
|
|
chr10_+_89267589
|
9.176
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr3_-_56809594
|
9.107
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr6_+_49467670
|
9.069
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr12_+_32832272
|
9.016
|
|
DNM1L
|
dynamin 1-like
|
|
chr5_+_141346401
|
8.924
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr15_+_42052574
|
8.784
|
|
MGA
|
MAX gene associated
|
|
chr4_+_57845057
|
8.736
|
|
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa
|
|
chr14_+_56078735
|
8.707
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr5_+_96038492
|
8.705
|
NM_173060
|
CAST
|
calpastatin
|
|
chr17_-_3599359
|
8.640
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr4_-_140223611
|
8.619
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr1_+_223889253
|
8.609
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr4_+_54292035
|
8.557
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr5_+_102465256
|
8.533
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr10_-_98336445
|
8.481
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_+_64231626
|
8.461
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr6_-_24877453
|
8.396
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr4_+_57845018
|
8.370
|
|
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa
|
|
chr10_-_91011516
|
8.349
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr1_+_144593626
|
8.332
|
|
NBPF10
|
neuroblastoma breakpoint family, member 10
|
|
chr4_+_146031528
|
8.307
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr17_-_38978824
|
8.291
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr7_+_7606634
|
8.289
|
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr1_+_148577356
|
8.121
|
NM_001102663
|
NBPF15
NBPF16
|
neuroblastoma breakpoint family, member 15
neuroblastoma breakpoint family, member 16
|
|
chr2_+_231281470
|
8.116
|
NM_001206704
|
SP100
|
SP100 nuclear antigen
|
|
chr9_-_36276930
|
8.112
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr19_-_46195171
|
8.094
|
|
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa
|
|
chr12_+_25205488
|
8.033
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr22_+_20909221
|
7.989
|
|
MED15
|
mediator complex subunit 15
|
|
chr10_-_91011562
|
7.979
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr14_-_99737821
|
7.940
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr12_+_69742140
|
7.932
|
|
LYZ
|
lysozyme
|
|
chr12_-_75905412
|
7.911
|
NM_007043
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr7_+_28725597
|
7.905
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr17_+_18768781
|
7.829
|
NM_001243940
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chr18_-_21017816
|
7.819
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chr5_-_150467220
|
7.801
|
NM_001252390
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr15_+_81489218
|
7.790
|
NM_001172128
|
IL16
|
interleukin 16
|
|
chr1_-_206785694
|
7.786
|
|
EIF2D
|
eukaryotic translation initiation factor 2D
|
|
chr1_+_167298382
|
7.752
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_+_49832214
|
7.750
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr11_+_87032428
|
7.711
|
|
TMEM135
|
transmembrane protein 135
|
|
chr2_+_15731769
|
7.706
|
NM_004939
|
DDX1
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 1
|
|
chr7_+_7606549
|
7.705
|
NM_019005
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr1_-_150738255
|
7.699
|
|
CTSS
|
cathepsin S
|
|
chr3_+_46395224
|
7.694
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|