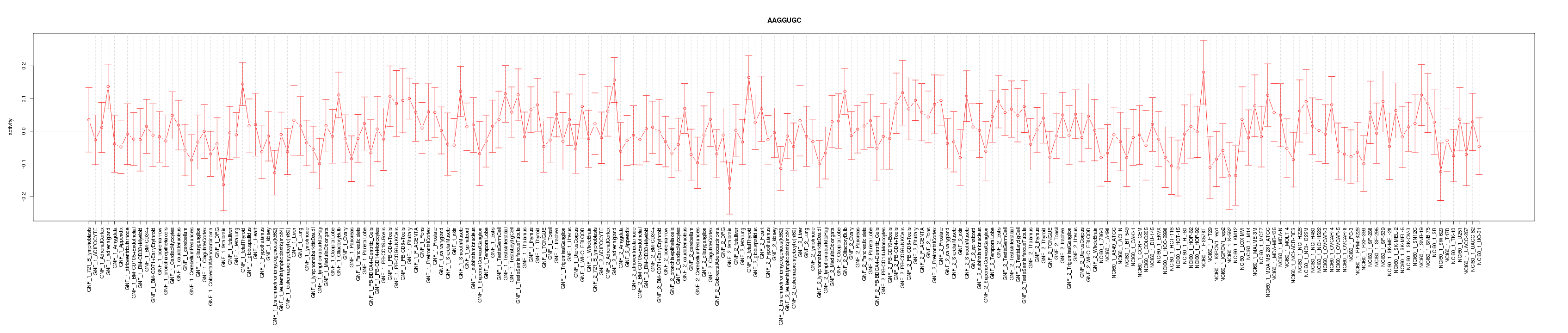
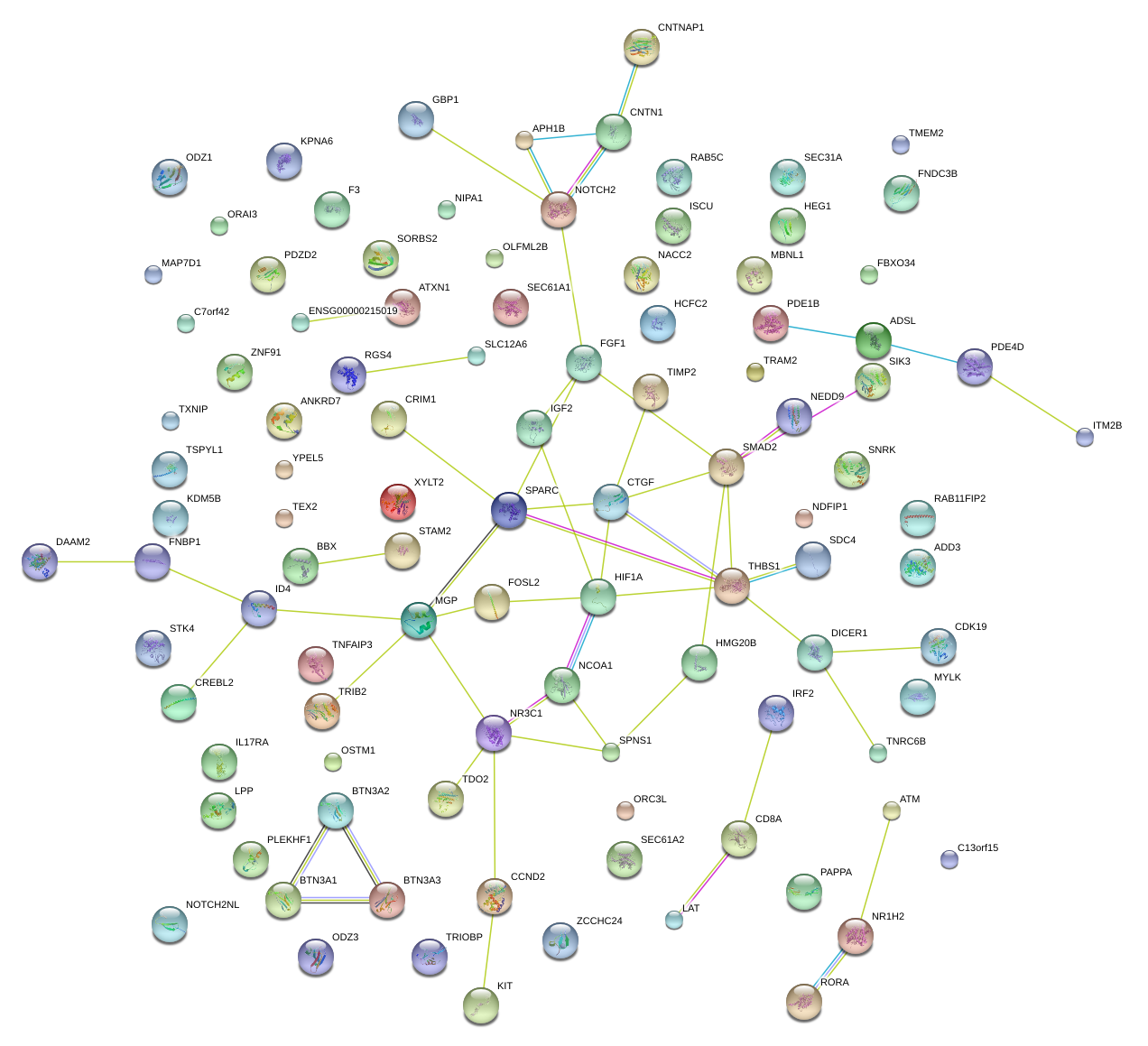
|
chr4_-_186877869
|
8.859
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732208
|
8.673
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186733372
|
8.629
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732047
|
7.918
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186697065
|
7.846
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_107241782
|
7.843
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr4_-_186578122
|
7.651
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_132272311
|
6.752
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr15_-_60919642
|
5.452
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_-_142815076
|
5.387
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr15_-_60884615
|
5.340
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_61521478
|
5.268
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_-_142783975
|
4.801
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr5_-_142783225
|
4.376
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr22_+_38092994
|
4.241
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr6_-_11382501
|
4.173
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_+_39760148
|
4.007
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr6_+_138188352
|
3.862
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_-_161993586
|
3.816
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr3_-_124774735
|
3.719
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr6_+_39760772
|
3.629
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr3_+_187871473
|
3.489
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_171757413
|
3.464
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr12_+_12764755
|
3.454
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr3_+_187943192
|
3.374
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_+_48423382
|
2.967
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr22_+_40573879
|
2.962
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_171758289
|
2.847
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr3_+_187930720
|
2.785
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_+_163038395
|
2.745
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_12856813
|
2.674
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr18_-_45457454
|
2.642
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr7_+_66386129
|
2.610
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr2_+_36583369
|
2.560
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr1_+_163041694
|
2.547
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr19_+_50879681
|
2.522
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr11_+_108093558
|
2.501
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr6_-_11232901
|
2.469
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr9_-_132805456
|
2.437
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr16_+_30960377
|
2.388
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr10_-_81205091
|
2.345
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_+_163038934
|
2.305
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038564
|
2.238
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_+_40440664
|
2.205
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr6_-_16761600
|
2.118
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr3_+_43328001
|
2.071
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr16_+_28996369
|
2.057
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr15_-_34610928
|
1.995
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_-_202777545
|
1.934
|
NM_006618
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr15_-_34630203
|
1.923
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr22_+_38142230
|
1.909
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr12_-_15038724
|
1.889
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr10_+_111767685
|
1.861
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr15_-_34629044
|
1.821
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr19_-_23578226
|
1.777
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr14_-_95608054
|
1.764
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr18_-_45456925
|
1.762
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr14_-_95623665
|
1.715
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_+_32573619
|
1.701
|
NM_012316
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7)
|
|
chr5_-_151066492
|
1.679
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr14_+_62162042
|
1.670
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr5_-_142077508
|
1.657
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_+_24807345
|
1.635
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr12_+_104458235
|
1.624
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr11_-_116968986
|
1.610
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr10_+_111765725
|
1.533
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr5_-_58334936
|
1.474
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_23086290
|
1.402
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr5_-_141993691
|
1.393
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_151985828
|
1.390
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr10_-_119805673
|
1.376
|
NM_014904
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chr14_-_95599794
|
1.240
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_+_30369749
|
1.205
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr1_+_145438437
|
1.185
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr17_+_40834571
|
1.147
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr15_+_39873276
|
1.147
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr3_-_123603144
|
1.129
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr12_+_4382882
|
1.120
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr3_+_127771126
|
1.115
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr15_-_34629941
|
1.089
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr9_-_74383301
|
1.063
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr1_+_36621751
|
1.057
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr16_+_28996146
|
1.049
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr11_-_2162340
|
1.046
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_+_152017193
|
1.025
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr1_-_120612301
|
1.024
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr20_+_43595119
|
1.022
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr5_+_141488314
|
0.988
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr11_-_2160199
|
0.978
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_-_123339423
|
0.971
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr5_-_142065586
|
0.968
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_142065991
|
0.955
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_62340652
|
0.952
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr11_-_2170792
|
0.950
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr13_+_42031525
|
0.948
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr7_+_117864711
|
0.946
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr9_+_118916069
|
0.920
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr5_+_31799030
|
0.913
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr9_-_138987096
|
0.909
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr2_-_87035518
|
0.899
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr6_-_108395939
|
0.883
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr19_+_3572903
|
0.879
|
NM_006339
|
HMG20B
|
high mobility group 20B
|
|
chr2_-_87018802
|
0.875
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr6_-_111136407
|
0.839
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_-_89530890
|
0.794
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr5_-_142000899
|
0.790
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_76921453
|
0.784
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr1_-_95007139
|
0.783
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr15_+_63569748
|
0.774
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr2_-_153032141
|
0.768
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr4_-_185395616
|
0.752
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr12_+_54943176
|
0.747
|
NM_000924
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr4_-_83812334
|
0.744
|
NM_001191049
|
SEC31A
|
SEC31 homolog A (S. cerevisiae)
|
|
chr20_-_43977023
|
0.731
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr4_+_55523906
|
0.695
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr14_+_55738871
|
0.689
|
NM_152231
|
FBXO34
|
F-box protein 34
|
|
chr6_+_26365397
|
0.683
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr13_+_48807250
|
0.683
|
NM_021999
|
ITM2B
|
integral membrane protein 2B
|
|
chr4_-_83812372
|
0.682
|
NM_001077206
NM_001077207
NM_001077208
NM_016211
NM_014933
|
SEC31A
|
SEC31 homolog A (S. cerevisiae)
|
|
chr6_-_116601102
|
0.676
|
NM_003309
|
TSPYL1
|
TSPY-like 1
|
|
chr6_-_52441815
|
0.634
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr2_+_28615771
|
0.630
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr12_+_108956293
|
0.622
|
NM_014301
NM_213595
|
ISCU
|
iron-sulfur cluster scaffold homolog (E. coli)
|
|
chr5_-_59783885
|
0.617
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_+_30156326
|
0.613
|
NM_024310
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr6_+_26368405
|
0.611
|
NM_001197246
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr6_+_26440699
|
0.610
|
NM_001242803
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr22_+_17565844
|
0.604
|
NM_014339
|
IL17RA
|
interleukin 17 receptor A
|
|
chr6_+_19837599
|
0.594
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr5_-_59064437
|
0.585
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_40307015
|
0.579
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr4_+_183245136
|
0.571
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr3_-_119812973
|
0.571
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr13_-_38172862
|
0.559
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr2_+_233561997
|
0.547
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr1_+_65886130
|
0.537
|
NM_001198683
NM_017526
|
LEPROT
|
leptin receptor overlapping transcript
|
|
chr8_-_18666295
|
0.532
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr18_+_47088400
|
0.522
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_+_220863627
|
0.520
|
NM_024709
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chr19_-_53606603
|
0.519
|
NM_001102603
NM_033288
NM_198893
|
ZNF160
|
zinc finger protein 160
|
|
chr6_-_35656677
|
0.463
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr14_+_70078309
|
0.461
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr1_+_25870043
|
0.431
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr7_-_139477437
|
0.426
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr1_-_182361311
|
0.425
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_153570270
|
0.422
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr22_-_20307511
|
0.417
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr4_-_2263706
|
0.406
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr3_+_9975513
|
0.405
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr11_+_94822937
|
0.403
|
NM_015036
|
ENDOD1
|
endonuclease domain containing 1
|
|
chr21_-_18985224
|
0.402
|
NM_001130914
NM_006806
|
BTG3
|
BTG family, member 3
|
|
chr4_+_2845552
|
0.401
|
NM_001119
NM_014189
NM_014190
NM_176801
|
ADD1
|
adducin 1 (alpha)
|
|
chr3_-_88199015
|
0.400
|
NM_001195308
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr1_+_21766559
|
0.381
|
NM_032264
|
NBPF3
|
neuroblastoma breakpoint family, member 3
|
|
chr3_+_108308336
|
0.378
|
NM_014648
|
DZIP3
|
DAZ interacting protein 3, zinc finger
|
|
chr2_-_97405799
|
0.378
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr18_-_3011944
|
0.373
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr10_-_101945686
|
0.369
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr1_+_173684061
|
0.367
|
NM_014458
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr14_-_20020271
|
0.363
|
NM_001145442
|
POTEG
POTEM
|
POTE ankyrin domain family, member G
POTE ankyrin domain family, member M
|
|
chr15_+_67358194
|
0.359
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_137071703
|
0.358
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr10_-_101945787
|
0.357
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr4_-_42658883
|
0.347
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr5_-_59189620
|
0.343
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_107168336
|
0.342
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr22_+_18893735
|
0.341
|
NM_005675
|
DGCR6
|
DiGeorge syndrome critical region gene 6
|
|
chr5_+_141346401
|
0.338
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr9_+_125703111
|
0.334
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr20_-_5093482
|
0.326
|
NM_001009923
NM_001009924
NM_001009925
NM_014145
|
C20orf30
|
chromosome 20 open reading frame 30
|
|
chr14_+_55738020
|
0.325
|
NM_017943
|
FBXO34
|
F-box protein 34
|
|
chr12_+_54955226
|
0.324
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr3_-_51533996
|
0.321
|
NM_001171904
NM_014703
|
VPRBP
|
Vpr (HIV-1) binding protein
|
|
chr21_+_45345278
|
0.317
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr7_+_90893735
|
0.311
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr20_+_43104513
|
0.310
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr14_-_23388380
|
0.305
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr15_+_81071708
|
0.305
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chrX_+_108780235
|
0.304
|
NM_001242618
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr6_+_52285804
|
0.300
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr1_+_117910052
|
0.299
|
NM_006699
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chrX_+_73641084
|
0.290
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr11_+_66886735
|
0.286
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chrX_+_108780058
|
0.283
|
NM_001242617
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr2_+_166428845
|
0.281
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr1_+_179712297
|
0.278
|
NM_173509
|
FAM163A
|
family with sequence similarity 163, member A
|
|
chr1_-_182360400
|
0.276
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr2_-_231090443
|
0.273
|
NM_001185015
|
SP110
|
SP110 nuclear body protein
|
|
chr19_+_4969123
|
0.272
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr13_+_111839172
|
0.270
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr9_+_115913227
|
0.267
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr2_-_202316227
|
0.265
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr6_+_52284948
|
0.262
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr2_+_202004997
|
0.257
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr3_+_183873253
|
0.247
|
NM_004423
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr3_-_13114547
|
0.241
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr4_+_175839508
|
0.241
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr9_+_96338671
|
0.232
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr5_+_141348692
|
0.231
|
NM_183401
|
RNF14
|
ring finger protein 14
|
|
chr15_+_67430358
|
0.227
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|