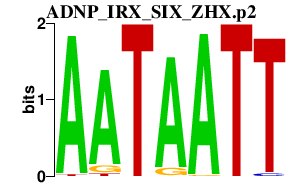
|
chr10_+_5136556
|
37.019
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_5136589
|
33.584
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_5136613
|
33.233
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr2_-_10942642
|
30.126
|
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr5_+_115898218
|
21.633
|
|
|
|
|
chr12_-_102512277
|
20.577
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr8_-_86290333
|
17.994
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr14_-_55650348
|
17.271
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr3_-_141747434
|
16.114
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr13_+_53035227
|
14.944
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr19_+_13134782
|
14.356
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr18_+_33234532
|
14.009
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr10_+_5005685
|
13.699
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr1_+_167298280
|
13.582
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_-_72649734
|
13.171
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr11_-_118900078
|
11.907
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr4_+_146031528
|
11.854
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr6_+_21593963
|
11.688
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr20_-_34243118
|
11.633
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr9_-_14086901
|
11.325
|
|
NFIB
|
nuclear factor I/B
|
|
chr12_+_20963637
|
10.639
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr15_-_56209305
|
10.452
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_+_64356430
|
10.390
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr10_+_5005601
|
10.341
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chrX_+_118714297
|
10.209
|
NM_181777
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr3_-_57234279
|
9.627
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr7_-_25268057
|
9.485
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr10_-_5045967
|
9.466
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr16_+_56817367
|
9.251
|
NM_001242796
|
NUP93
|
nucleoporin 93kDa
|
|
chr2_+_204055404
|
9.046
|
|
RPL12
|
ribosomal protein L12
|
|
chr7_-_26232148
|
8.449
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr11_+_114167197
|
8.402
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr12_+_50869531
|
8.208
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr11_+_114167168
|
8.077
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr6_+_34206567
|
7.988
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_+_89267589
|
7.969
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr11_+_114166534
|
7.633
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr1_+_68150858
|
7.502
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr3_-_113897849
|
7.418
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr12_+_54410893
|
7.396
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr4_+_140587086
|
7.314
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr1_+_74701070
|
7.289
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr10_-_75006963
|
7.279
|
NM_015190
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9
|
|
chr6_-_86351156
|
7.241
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_+_5005453
|
7.220
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr11_+_101983170
|
7.220
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr6_+_34204928
|
7.108
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_167298382
|
7.023
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_74269971
|
6.928
|
NM_000477
|
ALB
|
albumin
|
|
chr8_-_86253863
|
6.924
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr5_-_98262217
|
6.894
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr5_+_102465256
|
6.831
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chrX_+_99839789
|
6.817
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr9_+_97521930
|
6.735
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr1_+_236557643
|
6.662
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr4_-_7069801
|
6.580
|
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chrX_+_70765479
|
6.518
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr14_+_102276279
|
6.480
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chrX_+_123234462
|
6.459
|
|
STAG2
|
stromal antigen 2
|
|
chr6_+_64286348
|
6.368
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_-_100046262
|
6.347
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr1_+_64088886
|
6.342
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr17_-_64225496
|
6.235
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr22_-_36236421
|
6.187
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr8_-_72268720
|
6.043
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_-_70518964
|
5.934
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr2_+_234580504
|
5.905
|
NM_021027
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr6_+_49467670
|
5.883
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr17_-_46622274
|
5.818
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr8_+_9953061
|
5.715
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr17_-_15554809
|
5.625
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr14_-_69354900
|
5.599
|
|
ACTN1
|
actinin, alpha 1
|
|
chr2_+_234637772
|
5.591
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr2_-_190927321
|
5.566
|
NM_005259
|
MSTN
|
myostatin
|
|
chr13_+_53216564
|
5.514
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr6_-_138820580
|
5.475
|
|
|
|
|
chr9_-_104198045
|
5.447
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr9_-_86584079
|
5.442
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr7_+_115863004
|
5.362
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr19_-_40331153
|
5.285
|
|
FBL
|
fibrillarin
|
|
chr3_-_25683825
|
5.229
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr2_+_171784947
|
5.209
|
NM_001201428
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr7_-_27205148
|
5.196
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr6_+_136172802
|
5.175
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr14_+_58711457
|
5.159
|
NM_002788
NM_152132
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr11_-_27742224
|
5.098
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_+_58711599
|
5.088
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr3_+_63884074
|
5.069
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr7_-_72936538
|
4.990
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr5_+_138211084
|
4.863
|
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa
|
|
chr20_+_30326903
|
4.857
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr2_+_172821874
|
4.857
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr14_+_58711555
|
4.854
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr11_+_87032428
|
4.797
|
|
TMEM135
|
transmembrane protein 135
|
|
chr2_-_74382352
|
4.778
|
|
|
|
|
chr2_-_136634002
|
4.765
|
NM_005915
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr15_+_57511616
|
4.711
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr3_+_136649316
|
4.690
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr14_+_67831484
|
4.654
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr2_+_109204745
|
4.651
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_-_158656487
|
4.625
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr8_-_82395401
|
4.613
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr14_+_74035762
|
4.559
|
NM_006821
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr13_-_103719195
|
4.535
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr7_-_27205123
|
4.527
|
|
HOXA9
|
homeobox A9
|
|
chr4_+_1902352
|
4.519
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr4_-_74088773
|
4.465
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_-_38821286
|
4.457
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr2_-_133429069
|
4.438
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr5_+_65372744
|
4.429
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr5_+_44808967
|
4.428
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr15_+_49715374
|
4.390
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr3_+_151985828
|
4.365
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr8_+_66582107
|
4.349
|
NM_001145838
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chr12_-_54653369
|
4.340
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr17_-_39191100
|
4.327
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr12_-_102874340
|
4.289
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_+_162930230
|
4.284
|
NM_182796
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr10_+_75673346
|
4.277
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr11_+_113775517
|
4.276
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr2_+_228356287
|
4.262
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr21_-_33074101
|
4.241
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr12_-_14132920
|
4.238
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr17_-_49124128
|
4.234
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr2_-_61764661
|
4.194
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr2_+_109204904
|
4.139
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr11_+_99426872
|
4.114
|
NM_175566
|
CNTN5
|
contactin 5
|
|
chr5_+_6746100
|
4.086
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr15_+_94899163
|
4.073
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr12_+_16064105
|
4.037
|
NM_015954
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr5_-_111091947
|
4.032
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_179044835
|
4.023
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr6_-_36515245
|
4.001
|
NM_007271
|
STK38
|
serine/threonine kinase 38
|
|
chr20_+_32150139
|
3.987
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr12_-_91398757
|
3.981
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr3_-_196911001
|
3.965
|
NM_001204387
NM_001204388
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr9_+_135854097
|
3.964
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr5_-_140895407
|
3.962
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr6_+_167525294
|
3.948
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr15_+_52201590
|
3.946
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr18_-_51690988
|
3.924
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr3_+_149192367
|
3.921
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr21_-_36421586
|
3.918
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_234545099
|
3.903
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr16_+_56815703
|
3.896
|
NM_001242795
|
NUP93
|
nucleoporin 93kDa
|
|
chr2_+_216986817
|
3.877
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chrX_-_55057408
|
3.851
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr4_-_7069755
|
3.840
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr10_+_94352960
|
3.822
|
|
KIF11
|
kinesin family member 11
|
|
chr14_+_102276125
|
3.804
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chrX_-_18690187
|
3.755
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr1_+_78470593
|
3.735
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr1_-_93257950
|
3.730
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr14_-_23504353
|
3.650
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr12_-_54653317
|
3.625
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr22_-_37098620
|
3.569
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr10_-_98325100
|
3.545
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr3_-_27498244
|
3.536
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr1_-_159684274
|
3.528
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr6_-_155777036
|
3.526
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr2_-_200329810
|
3.518
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr1_+_32661018
|
3.508
|
|
TXLNA
|
taxilin alpha
|
|
chr7_+_107110501
|
3.506
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr2_+_211458078
|
3.471
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr5_+_132432880
|
3.447
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr4_+_155484131
|
3.446
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr20_-_7921068
|
3.441
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr6_-_25874439
|
3.419
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr6_+_34204576
|
3.388
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_+_149192479
|
3.368
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr17_-_46682333
|
3.348
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr4_-_111563278
|
3.343
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr17_-_79876024
|
3.337
|
|
SIRT7
|
sirtuin 7
|
|
chr3_-_194188967
|
3.335
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr2_+_109223600
|
3.331
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr4_+_41614725
|
3.322
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_168250277
|
3.318
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr11_-_83393436
|
3.315
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr13_+_98605928
|
3.283
|
NM_002271
|
IPO5
|
importin 5
|
|
chr2_+_86333304
|
3.265
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chr17_-_48785016
|
3.265
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr9_+_36572844
|
3.225
|
NM_014791
|
MELK
|
maternal embryonic leucine zipper kinase
|
|
chr8_-_6420454
|
3.201
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr7_+_28452143
|
3.199
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr14_-_65409445
|
3.190
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr4_+_54292035
|
3.187
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr10_-_94257470
|
3.180
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr5_-_95158465
|
3.173
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr9_+_79968344
|
3.171
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr22_-_36236264
|
3.162
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_+_155538096
|
3.148
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr7_+_80275634
|
3.143
|
NM_001127443
NM_001127444
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr15_+_66797632
|
3.132
|
|
ZWILCH
|
Zwilch, kinetochore associated, homolog (Drosophila)
|
|
chr6_-_75965845
|
3.127
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr2_-_192711923
|
3.125
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr5_+_126859157
|
3.108
|
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr2_+_234627423
|
3.064
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr7_+_134212343
|
3.047
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr5_-_95158365
|
3.041
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr17_+_28268622
|
3.018
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|