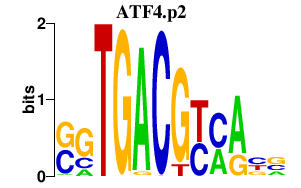
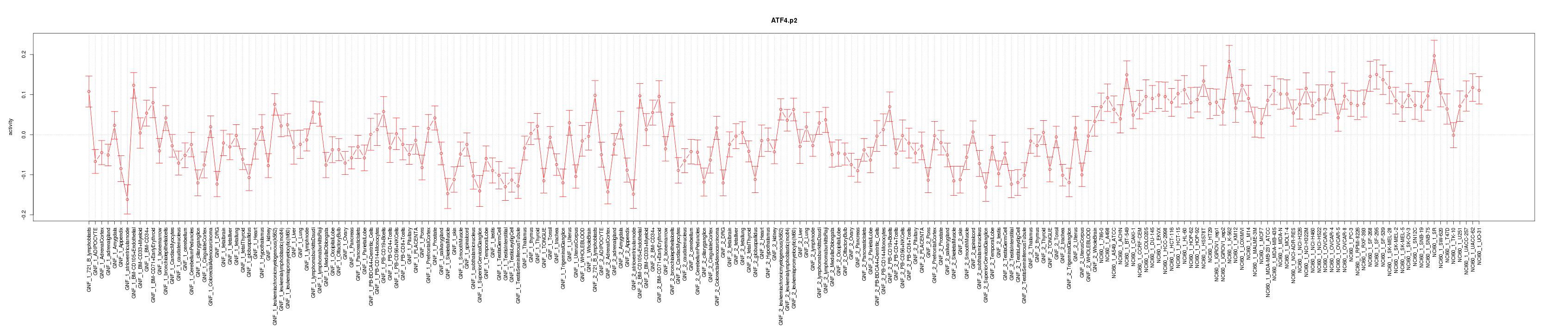
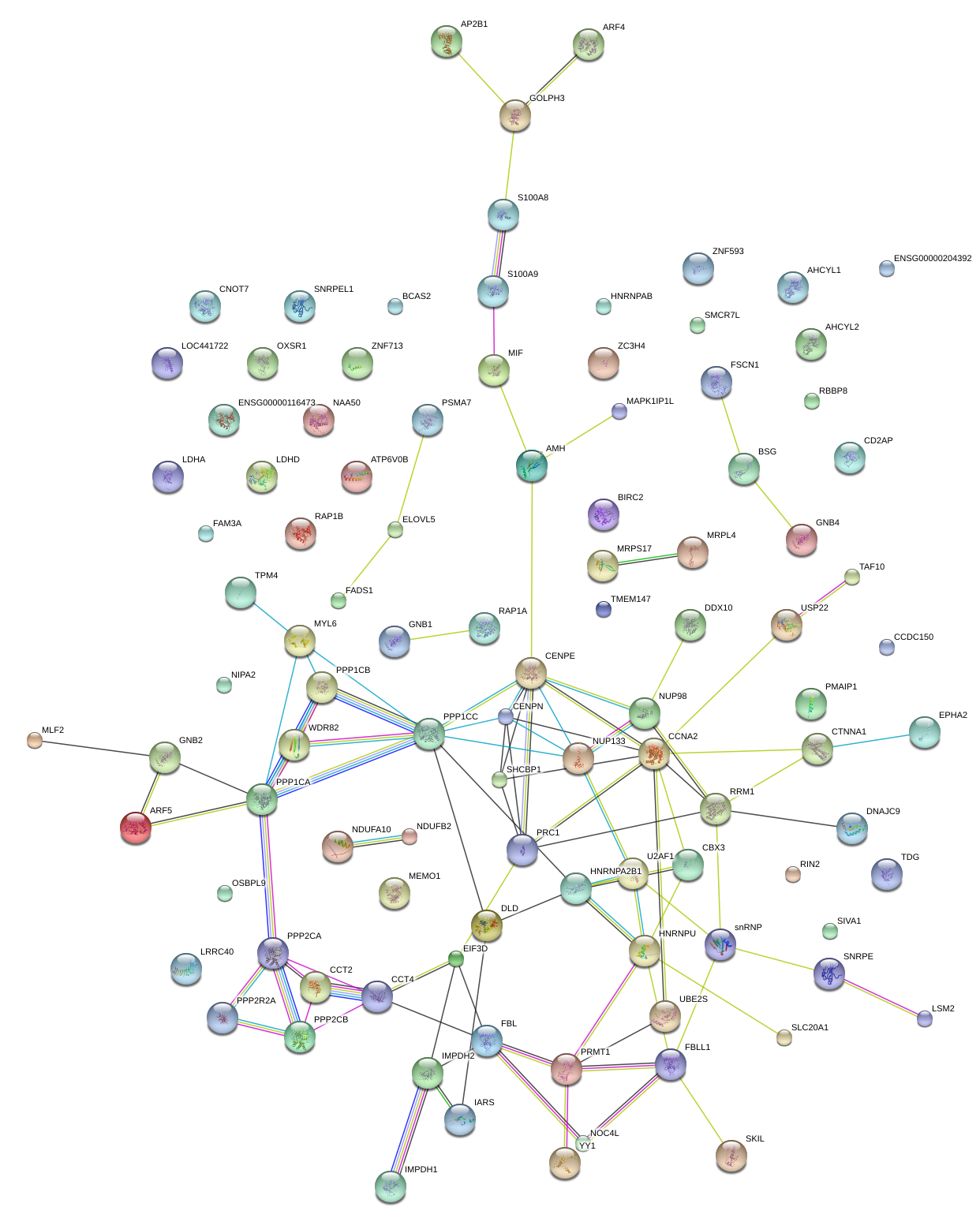
|
chr12_-_123380608
|
91.173
|
|
|
|
|
chr5_-_133561745
|
70.025
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr19_+_16187306
|
69.868
|
|
TPM4
|
tropomyosin 4
|
|
chr8_-_17104266
|
68.307
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr6_+_47445440
|
62.788
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr11_+_4116059
|
61.361
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr22_+_24236579
|
60.840
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr22_+_24236575
|
60.029
|
|
|
|
|
chr22_+_24236569
|
59.882
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr19_+_16187266
|
59.790
|
|
TPM4
|
tropomyosin 4
|
|
chr19_-_55918980
|
57.148
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr22_+_24236586
|
57.041
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr22_+_24236556
|
54.700
|
NM_002415
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr1_+_26496382
|
54.658
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr19_+_16187315
|
52.173
|
|
TPM4
|
tropomyosin 4
|
|
chr8_+_26149004
|
50.614
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr8_+_26149043
|
49.522
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr11_+_4115923
|
48.082
|
NM_001033
|
RRM1
|
ribonucleotide reductase M1
|
|
chr22_+_24236595
|
46.073
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr5_-_133561551
|
45.987
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr19_-_47617008
|
44.738
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr8_-_17104103
|
44.223
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr11_+_4116077
|
43.109
|
|
RRM1
|
ribonucleotide reductase M1
|
|
chr2_-_32236104
|
42.375
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr1_-_70671263
|
40.860
|
NM_017768
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr3_+_38206968
|
40.799
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr3_-_49066760
|
39.963
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr15_-_91537724
|
39.708
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr3_-_49066858
|
39.086
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr7_+_56019552
|
38.650
|
NM_015969
|
MRPS17
|
mitochondrial ribosomal protein S17
|
|
chr1_+_52082755
|
38.366
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr6_-_31774579
|
37.749
|
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_-_245025958
|
37.127
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr2_-_240964717
|
37.029
|
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr12_-_6862131
|
36.219
|
|
MLF2
|
myeloid leukemia factor 2
|
|
chr18_+_20513277
|
35.628
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr3_-_49066813
|
35.420
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr6_-_53213586
|
35.240
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr4_-_122744978
|
34.562
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr4_-_104119475
|
33.995
|
NM_001813
|
CENPE
|
centromere protein E, 312kDa
|
|
chr4_-_122744782
|
33.863
|
|
CCNA2
|
cyclin A2
|
|
chr11_+_102217947
|
33.742
|
NM_001166
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr8_+_26149080
|
33.539
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr7_+_140396463
|
33.253
|
NM_004546
|
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa
|
|
chr2_+_113403433
|
33.249
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr19_+_50180408
|
33.118
|
NM_001207042
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr1_-_115124211
|
33.049
|
NM_005872
|
BCAS2
|
breast carcinoma amplified sequence 2
|
|
chr19_+_50180492
|
32.730
|
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr12_+_69979268
|
32.466
|
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr15_-_23034307
|
32.442
|
|
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2
|
|
chr6_-_31774725
|
32.432
|
NM_021177
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr7_+_100273776
|
31.559
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr19_-_40337018
|
31.441
|
NM_001436
|
FBL
|
fibrillarin
|
|
chr7_+_107531585
|
30.961
|
NM_000108
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr11_-_67188614
|
30.567
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr10_-_75006963
|
30.517
|
NM_015190
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9
|
|
chr1_-_70671160
|
30.509
|
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr14_+_105219466
|
30.091
|
NM_006427
NM_021709
|
SIVA1
|
SIVA1, apoptosis-inducing factor
|
|
chr19_-_40336944
|
29.885
|
|
FBL
|
fibrillarin
|
|
chr12_+_69979202
|
29.871
|
NM_006431
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr2_-_240964807
|
29.633
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr7_+_107531618
|
29.402
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr7_+_107531616
|
29.121
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr3_-_52312655
|
28.894
|
NM_025222
|
WDR82
|
WD repeat domain 82
|
|
chr11_-_61584451
|
28.795
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr5_+_10353980
|
28.303
|
|
|
|
|
chr5_-_32174385
|
27.892
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr14_+_100705101
|
27.868
|
NM_003403
|
YY1
|
YY1 transcription factor
|
|
chr20_+_19915716
|
27.529
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr2_-_62115714
|
27.138
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr7_+_26240830
|
27.027
|
NM_007276
|
CBX3
|
chromobox homolog 3
|
|
chr5_+_138211084
|
26.968
|
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa
|
|
chr12_+_69979447
|
26.726
|
NM_001198842
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr22_+_39898365
|
26.533
|
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr1_+_203830723
|
26.412
|
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr2_-_62115726
|
26.363
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr1_+_110527327
|
26.331
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr11_+_108535773
|
26.256
|
NM_004398
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10
|
|
chr12_+_69004684
|
26.218
|
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr17_+_33914278
|
25.873
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr9_-_95055913
|
25.711
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr19_+_16187047
|
25.653
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr5_-_32174397
|
25.510
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr5_-_32174355
|
25.454
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr19_-_40336997
|
25.067
|
|
FBL
|
fibrillarin
|
|
chr19_+_36036448
|
24.828
|
NM_001242597
NM_001242598
NM_032635
|
TMEM147
|
transmembrane protein 147
|
|
chr16_+_81040847
|
24.532
|
|
CENPN
|
centromere protein N
|
|
chr11_-_6633400
|
24.252
|
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr1_+_203830732
|
24.249
|
NM_003094
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr21_-_44527603
|
24.127
|
|
U2AF1
|
U2 small nuclear RNA auxiliary factor 1
|
|
chr12_+_69004710
|
23.992
|
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr1_+_44440623
|
23.812
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr14_+_55518398
|
23.687
|
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr12_+_132629014
|
23.647
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr12_+_104359557
|
23.580
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr9_-_95055966
|
23.475
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr11_+_18416111
|
23.300
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr1_-_229644077
|
23.281
|
NM_018230
|
NUP133
|
nucleoporin 133kDa
|
|
chr18_+_57567191
|
23.119
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr1_+_44440595
|
23.056
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr7_-_26240364
|
23.046
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr12_+_56552141
|
23.044
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr11_+_18416106
|
23.027
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr5_+_177631533
|
22.584
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr3_+_170075521
|
22.527
|
|
SKIL
|
SKI-like oncogene
|
|
chr2_-_32235603
|
22.506
|
NM_001137602
NM_015955
|
MEMO1
|
mediator of cell motility 1
|
|
chr22_-_36925212
|
22.425
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr1_+_44440642
|
22.358
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr2_-_32235722
|
22.335
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr14_+_55518313
|
22.313
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr19_+_572568
|
22.174
|
|
BSG
|
basigin (Ok blood group)
|
|
chr9_-_95055986
|
22.136
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr7_+_127228405
|
22.128
|
NM_001662
|
ARF5
|
ADP-ribosylation factor 5
|
|
chr1_-_16482426
|
22.087
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr7_+_5632435
|
22.038
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr19_+_10362912
|
22.031
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr3_-_113465101
|
21.963
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr2_-_62115744
|
21.878
|
NM_006430
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr17_-_20946088
|
21.786
|
NM_015276
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr20_-_60718440
|
21.597
|
NM_002792
|
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7
|
|
chr22_-_36925270
|
21.573
|
NM_003753
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr11_-_3818971
|
21.550
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chr1_-_229643996
|
21.526
|
|
NUP133
|
nucleoporin 133kDa
|
|
chr16_-_46655303
|
21.464
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr14_+_55518372
|
21.392
|
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr5_-_32174327
|
21.362
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr22_+_39898248
|
21.307
|
NM_019008
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr7_+_44646220
|
21.278
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr7_-_26240320
|
21.253
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr2_+_242255365
|
21.174
|
|
SEPT2
|
septin 2
|
|
chr7_-_105925329
|
21.146
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr11_+_95523608
|
21.145
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr7_-_26240366
|
21.110
|
NM_002137
NM_031243
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr18_+_158586
|
21.064
|
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr22_-_36925138
|
21.042
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr19_-_14529853
|
20.974
|
|
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
|
|
chr14_-_103523619
|
20.968
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|
|
chr2_-_62115641
|
20.879
|
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta)
|
|
chr1_+_70671331
|
20.795
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr8_+_98788014
|
20.705
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr6_-_3157696
|
20.662
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A class IIa
|
|
chr3_+_127317274
|
20.628
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr19_-_8070468
|
20.282
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr17_+_80674579
|
20.278
|
NM_024619
|
FN3KRP
|
fructosamine 3 kinase related protein
|
|
chr17_+_15902693
|
20.088
|
NM_017775
|
TTC19
|
tetratricopeptide repeat domain 19
|
|
chr3_+_38207247
|
20.074
|
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr11_-_28129646
|
19.957
|
|
KIF18A
|
kinesin family member 18A
|
|
chr1_+_203830763
|
19.906
|
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr19_+_10362886
|
19.880
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr2_-_10588451
|
19.866
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr19_-_14530129
|
19.785
|
|
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
|
|
chr3_-_133380626
|
19.627
|
NM_007027
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1
|
|
chr1_-_33283632
|
19.598
|
NM_003680
|
YARS
|
tyrosyl-tRNA synthetase
|
|
chr12_+_54378929
|
19.564
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr5_-_133561805
|
19.454
|
NM_002715
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chrX_+_153672490
|
19.441
|
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr17_-_56084611
|
19.433
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr19_-_55919324
|
19.302
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr22_+_38597938
|
19.301
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr3_-_156877882
|
19.296
|
|
CCNL1
|
cyclin L1
|
|
chr5_-_133304274
|
19.267
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr1_-_31769599
|
19.227
|
NM_004814
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5)
|
|
chr4_+_170541690
|
19.190
|
|
CLCN3
|
chloride channel 3
|
|
chr11_+_118230348
|
19.095
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr2_-_70520780
|
18.996
|
|
SNRPG
|
small nuclear ribonucleoprotein polypeptide G
|
|
chr18_+_158480
|
18.987
|
NM_001037334
NM_005151
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr19_+_1941157
|
18.973
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr1_-_32110464
|
18.942
|
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr19_+_13263890
|
18.822
|
|
IER2
|
immediate early response 2
|
|
chr16_+_28722977
|
18.807
|
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr19_+_36630999
|
18.769
|
|
CAPNS1
|
calpain, small subunit 1
|
|
chr16_-_46655262
|
18.715
|
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr19_+_572575
|
18.680
|
|
BSG
|
basigin (Ok blood group)
|
|
chrX_+_153672449
|
18.624
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr11_-_6633428
|
18.508
|
NM_006284
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chrX_+_67718868
|
18.463
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr19_+_572403
|
18.431
|
NM_001728
NM_198589
|
BSG
|
basigin (Ok blood group)
|
|
chr9_-_127952037
|
18.424
|
NM_001123355
NM_001123369
NM_002721
|
PPP6C
|
protein phosphatase 6, catalytic subunit
|
|
chr5_+_177631534
|
18.386
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr17_+_8152594
|
18.383
|
NM_012393
|
PFAS
|
phosphoribosylformylglycinamidine synthase
|
|
chr22_-_36925175
|
18.339
|
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr14_+_100705279
|
18.291
|
|
YY1
|
YY1 transcription factor
|
|
chr1_+_110527432
|
18.290
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr10_+_85899184
|
18.210
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr19_-_59066477
|
18.194
|
NM_014453
|
CHMP2A
|
charged multivesicular body protein 2A
|
|
chr8_-_146017774
|
18.178
|
|
RPL8
|
ribosomal protein L8
|
|
chr3_+_128445026
|
18.153
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr9_-_111696173
|
18.075
|
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein
|
|
chr22_+_32340435
|
18.026
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr4_+_17812524
|
17.997
|
NM_022346
|
NCAPG
|
non-SMC condensin I complex, subunit G
|
|
chr11_-_6633178
|
17.993
|
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr5_-_32174242
|
17.912
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr18_+_72163335
|
17.830
|
NM_018235
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr16_-_3767449
|
17.771
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr20_+_361675
|
17.751
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr1_-_53704121
|
17.689
|
NM_002370
|
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila)
|
|
chr6_-_42713837
|
17.649
|
NM_003192
|
TBCC
|
tubulin folding cofactor C
|
|
chr19_+_36631001
|
17.550
|
|
CAPNS1
|
calpain, small subunit 1
|
|
chr10_-_33247131
|
17.459
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_+_44440673
|
17.396
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|