|
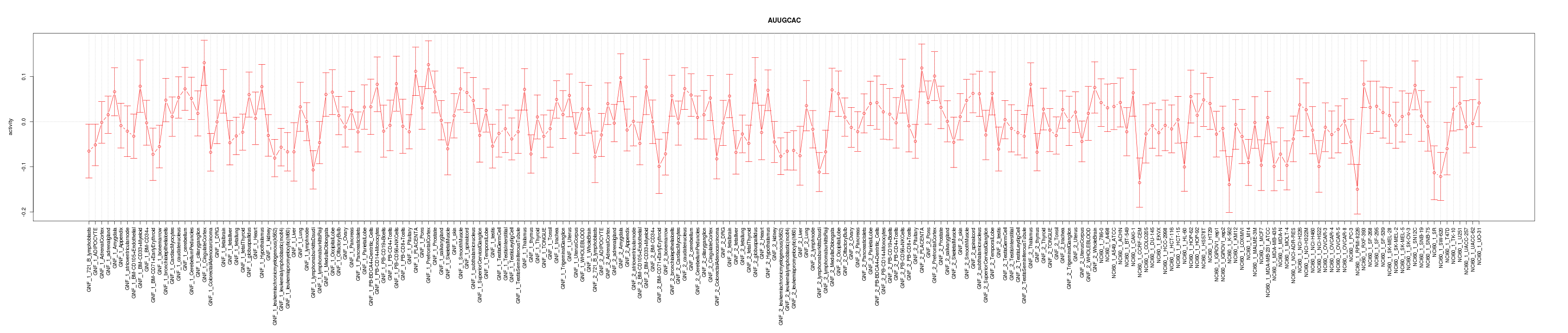
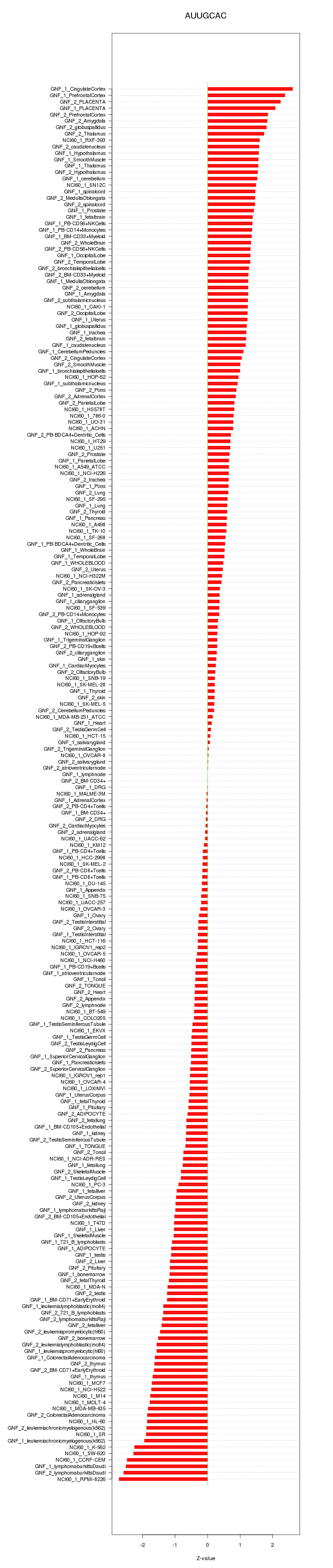
chr8_-_24814028
|
24.796
|
NM_006158
|
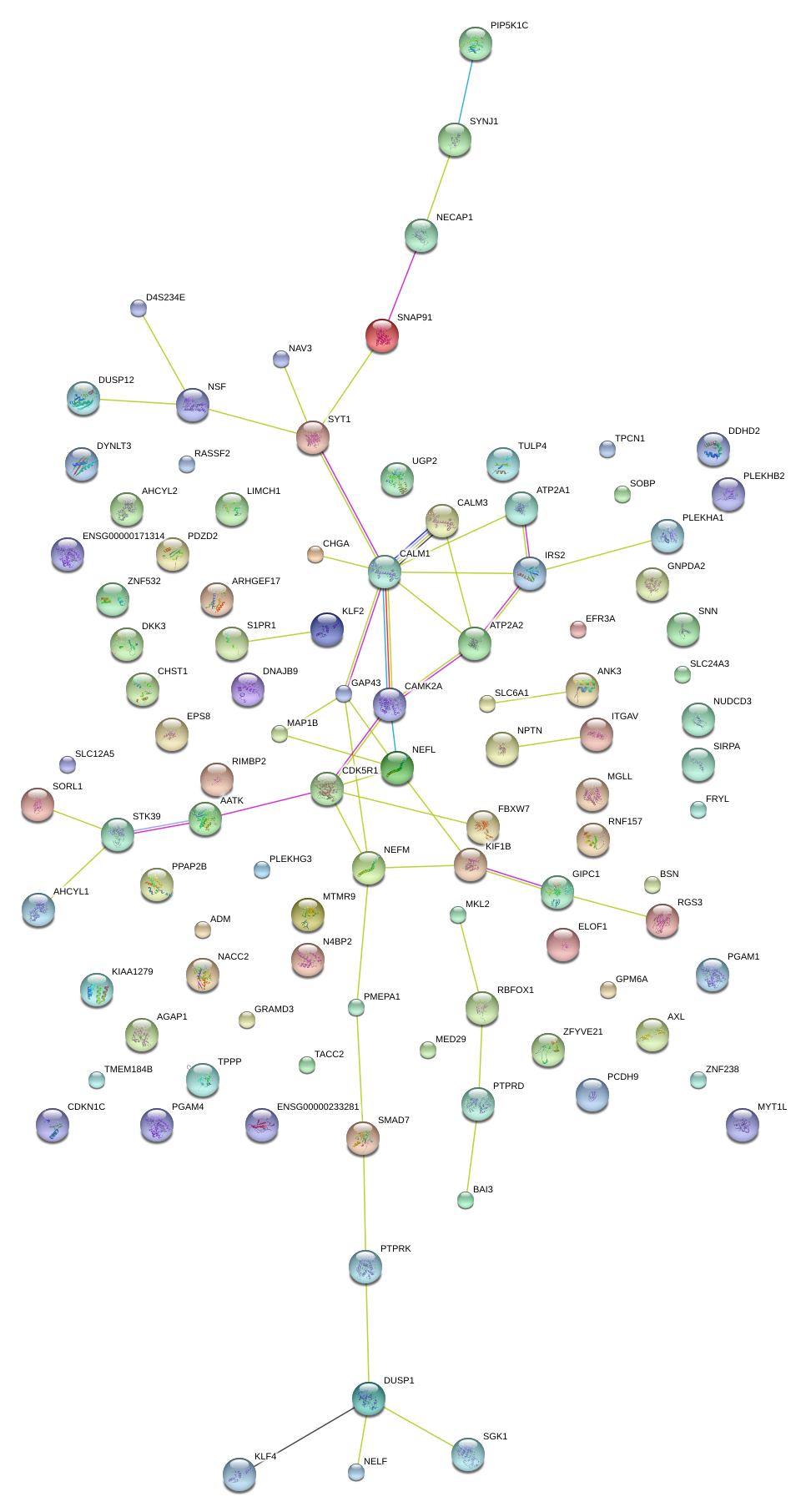
NEFL
|
neurofilament, light polypeptide
|
|
chr20_+_44650260
|
17.968
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr20_+_44657812
|
17.783
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr8_+_24772454
|
16.327
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_24771268
|
16.007
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr11_-_12030882
|
14.659
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_12030128
|
14.345
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_12030622
|
14.055
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr17_+_44668034
|
13.949
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr4_-_176923572
|
13.880
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_176734181
|
13.830
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr5_+_71403040
|
12.468
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_127541975
|
11.804
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr12_+_79257772
|
11.780
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr9_-_138987096
|
11.770
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr12_+_79439432
|
11.411
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr3_+_115342150
|
11.263
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr6_-_84419108
|
10.894
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr12_+_79258448
|
10.752
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr6_-_84418705
|
10.732
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr3_-_127541160
|
10.422
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr13_-_67804432
|
10.204
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr5_+_125758818
|
10.187
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr13_-_110438896
|
9.944
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr4_+_41362750
|
9.594
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_+_125800786
|
9.494
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr19_-_3700466
|
9.482
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr6_+_107811272
|
9.210
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_+_31799030
|
9.200
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr4_+_41614911
|
9.175
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_+_125695787
|
8.982
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr1_+_110543548
|
8.980
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chrX_-_37706816
|
8.916
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr15_-_73925658
|
8.798
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr2_+_187464931
|
8.786
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr16_+_11762235
|
8.757
|
NM_003498
|
SNN
|
stannin
|
|
chr6_-_134496006
|
8.651
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_11034419
|
8.510
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr6_-_134639195
|
8.429
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_+_8234796
|
8.334
|
NM_015509
|
NECAP1
|
NECAP endocytosis associated 1
|
|
chr6_-_134498973
|
8.258
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_110527327
|
8.205
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_-_79105747
|
8.100
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr6_-_134497069
|
8.073
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_2906960
|
7.959
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr1_+_110546569
|
7.948
|
NM_001242675
NM_001242676
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr8_+_132916339
|
7.797
|
NM_015137
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr10_+_124134093
|
7.744
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr2_+_187454724
|
7.659
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr16_+_6069094
|
7.616
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_-_79139871
|
7.354
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr11_+_121322848
|
7.293
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr16_+_6823809
|
7.290
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr10_+_124145584
|
7.072
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr20_+_1874779
|
7.019
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr1_+_10270680
|
6.954
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr20_+_1875825
|
6.925
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr6_+_158733691
|
6.903
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr14_+_93389444
|
6.900
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr4_-_153303663
|
6.807
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr22_-_38668966
|
6.800
|
NM_001195072
NM_012264
|
TMEM184B
|
transmembrane protein 184B
|
|
chr22_-_38668669
|
6.718
|
NM_001195071
|
TMEM184B
|
transmembrane protein 184B
|
|
chr3_+_49591897
|
6.697
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr7_+_108210188
|
6.332
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr14_+_65171149
|
6.244
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr20_+_1875382
|
6.160
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr4_-_153456152
|
6.138
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr16_+_7382750
|
6.078
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr19_+_47104490
|
6.023
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr5_-_149669400
|
5.947
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_+_131862419
|
5.904
|
NM_001100623
NM_017958
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2
|
|
chr10_+_124151818
|
5.883
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr2_-_2334887
|
5.840
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr5_-_693413
|
5.795
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr6_-_128841432
|
5.705
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr18_+_56530060
|
5.694
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr2_+_236402685
|
5.653
|
NM_001037131
NM_001244888
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr7_-_44530178
|
5.596
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr9_-_110252045
|
5.581
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_+_10326618
|
5.508
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr17_+_30813928
|
5.445
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr12_+_110719029
|
5.428
|
NM_001681
NM_170665
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr8_+_38089165
|
5.270
|
NM_001164232
|
DDHD2
|
DDHD domain containing 2
|
|
chr10_+_70748476
|
5.194
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr20_-_4795768
|
5.158
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr19_+_16435632
|
5.140
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_+_73019662
|
4.869
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr19_-_14606939
|
4.840
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr20_-_4804067
|
4.837
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr10_+_99186017
|
4.791
|
NM_002629
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr17_-_74236272
|
4.745
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr21_-_34100316
|
4.690
|
NM_003895
NM_203446
|
SYNJ1
|
synaptojanin 1
|
|
chr2_+_64068074
|
4.618
|
NM_001001521
|
UGP2
|
UDP-glucose pyrophosphorylase 2
|
|
chr9_-_10612722
|
4.468
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr14_+_104182045
|
4.399
|
NM_001198953
NM_024071
|
ZFYVE21
|
zinc finger, FYVE domain containing 21
|
|
chr1_+_244214552
|
4.372
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr18_-_46475702
|
4.356
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr20_+_19193282
|
4.336
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr6_+_69344939
|
4.281
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr4_+_4387982
|
4.259
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr12_+_113659216
|
4.230
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr16_+_14164879
|
4.224
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr12_+_78225068
|
4.197
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr11_-_45687135
|
4.144
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr9_+_116207010
|
4.106
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_+_64068984
|
4.099
|
NM_006759
|
UGP2
|
UDP-glucose pyrophosphorylase 2
|
|
chr12_-_131002409
|
4.081
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr12_-_15942350
|
4.069
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_+_123748688
|
4.053
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr22_-_36236421
|
3.976
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr10_-_62493282
|
3.918
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_39881950
|
3.852
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr9_-_140353785
|
3.831
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr2_-_169103886
|
3.799
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr10_-_62149487
|
3.789
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_41725098
|
3.781
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr22_-_36424397
|
3.748
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_48782281
|
3.745
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr5_-_172198160
|
3.718
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr20_-_56286540
|
3.699
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr4_-_44728518
|
3.671
|
NM_138335
|
GNPDA2
|
glucosamine-6-phosphate deaminase 2
|
|
chr8_+_11141983
|
3.646
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr4_+_40058507
|
3.636
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr1_-_57045235
|
3.629
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr4_-_153273683
|
3.622
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_+_101702304
|
3.597
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr5_+_109025135
|
3.585
|
NM_002372
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr1_+_51701943
|
3.545
|
NM_014372
|
RNF11
|
ring finger protein 11
|
|
chr1_-_94703252
|
3.533
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr10_+_1102741
|
3.524
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr15_-_52861177
|
3.502
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr9_+_116327301
|
3.492
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116342939
|
3.486
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr17_+_11924116
|
3.473
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr11_-_790103
|
3.445
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr7_-_35734762
|
3.445
|
NM_022373
|
HERPUD2
|
HERPUD family member 2
|
|
chr8_+_79578258
|
3.439
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr18_-_21166440
|
3.406
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr3_-_120169493
|
3.388
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr3_+_4535030
|
3.356
|
NM_001099952
NM_001168272
NM_002222
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr4_-_40517913
|
3.352
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr9_+_116356199
|
3.315
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_31381436
|
3.309
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr1_-_27481400
|
3.300
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr20_+_24449834
|
3.277
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr13_-_77901176
|
3.254
|
NM_015057
|
MYCBP2
|
MYC binding protein 2
|
|
chr22_+_29876180
|
3.236
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr7_+_94138991
|
3.210
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr6_-_16761600
|
3.160
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr17_-_42580927
|
3.102
|
NM_001002909
|
GPATCH8
|
G patch domain containing 8
|
|
chr6_-_88875766
|
3.068
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr1_+_244216506
|
3.056
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr17_-_1466024
|
2.933
|
NM_006224
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr21_-_34100228
|
2.909
|
NM_001160302
NM_001160306
|
SYNJ1
|
synaptojanin 1
|
|
chr13_+_32605436
|
2.895
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr10_+_96162185
|
2.895
|
NM_015188
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr5_-_179780314
|
2.887
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr8_+_58906968
|
2.835
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr18_-_46477080
|
2.811
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr18_-_46469118
|
2.803
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_24807345
|
2.792
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr3_+_171758289
|
2.778
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr22_-_29075708
|
2.706
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr17_-_53499055
|
2.685
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr3_+_171757413
|
2.653
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr2_+_26915390
|
2.643
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr1_+_180601121
|
2.637
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr8_+_38089008
|
2.600
|
NM_001164234
NM_015214
|
DDHD2
|
DDHD domain containing 2
|
|
chr18_-_53255734
|
2.575
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr3_-_15106806
|
2.575
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr12_-_65153189
|
2.557
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr17_-_8066254
|
2.549
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr10_-_62332666
|
2.533
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr9_-_14398981
|
2.532
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr19_+_40973049
|
2.503
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr12_-_133405197
|
2.474
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr9_+_134305476
|
2.420
|
NM_013318
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr6_-_80656870
|
2.415
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr10_-_73848766
|
2.393
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chrX_+_70586113
|
2.393
|
NM_004606
NM_138923
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa
|
|
chr1_-_114354906
|
2.386
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr3_+_123813527
|
2.368
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr1_-_202679415
|
2.322
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr20_+_30865435
|
2.316
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr1_+_183605207
|
2.294
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr1_-_202612580
|
2.279
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr15_-_83316621
|
2.278
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr4_+_169418167
|
2.273
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr7_-_105029321
|
2.249
|
NM_182692
|
SRPK2
|
SRSF protein kinase 2
|
|
chr4_+_169753105
|
2.246
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr3_+_23958294
|
2.134
|
NM_001253379
NM_001253384
NM_002948
|
RPL15
|
ribosomal protein L15
|
|
chr13_+_48807250
|
2.109
|
NM_021999
|
ITM2B
|
integral membrane protein 2B
|
|
chr10_+_64564480
|
2.101
|
NM_032804
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase
|
|
chr4_+_169552752
|
2.085
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr8_+_77593506
|
2.034
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr8_+_41348067
|
2.009
|
NM_001174124
NM_016099
|
GOLGA7
|
golgin A7
|
|
chr15_-_28567249
|
2.003
|
NM_004667
|
HERC2
|
hect domain and RLD 2
|
|
chr14_+_59104736
|
2.002
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr1_-_221915458
|
1.986
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr14_+_92790151
|
1.984
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|