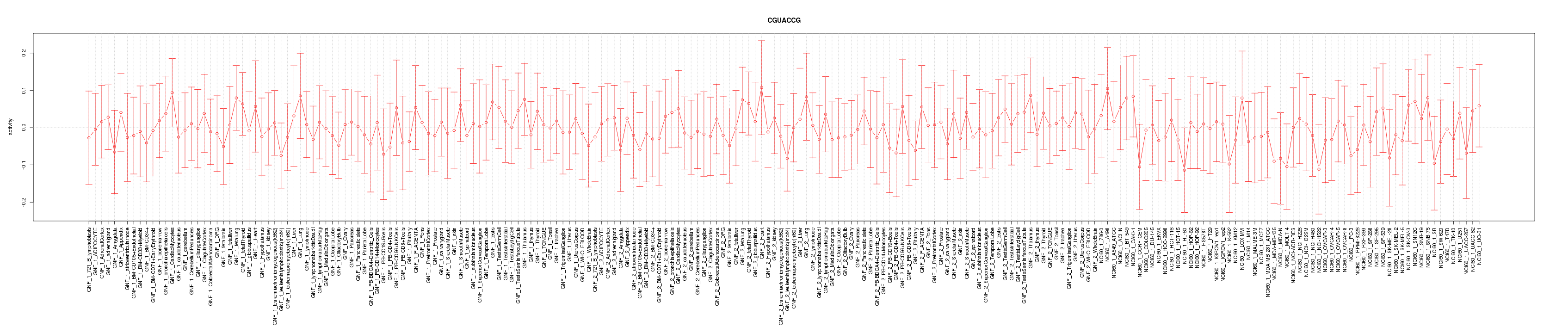
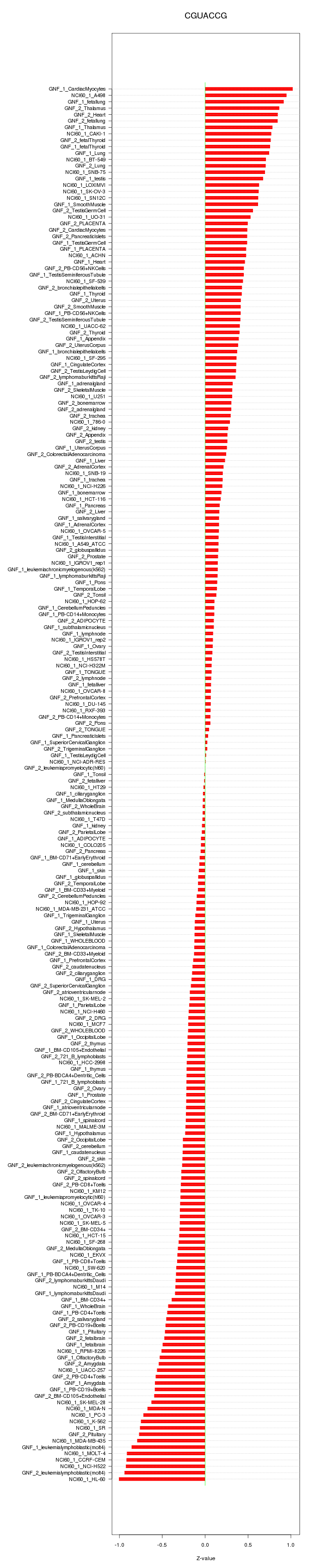
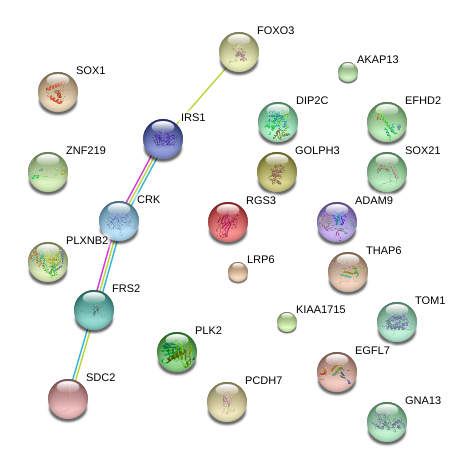
|
11.55
|
2.28e-03
|
GO:0007243
|
intracellular protein kinase cascade
|
|
11.55
|
2.28e-03
|
GO:0023014
|
signal transduction via phosphorylation event
|
|
5.25
|
4.89e-03
|
GO:0009966
|
regulation of signal transduction
|
|
9.92
|
5.42e-03
|
GO:0043549
|
regulation of kinase activity
|
|
9.58
|
6.62e-03
|
GO:0051338
|
regulation of transferase activity
|
|
22.46
|
7.11e-03
|
GO:0008286
|
insulin receptor signaling pathway
|
|
47.93
|
8.75e-03
|
GO:0000186
|
activation of MAPKK activity
|
|
4.57
|
1.49e-02
|
GO:0023051
|
regulation of signaling
|
|
17.32
|
1.97e-02
|
GO:0032869
|
cellular response to insulin stimulus
|
|
7.55
|
2.53e-02
|
GO:0007167
|
enzyme linked receptor protein signaling pathway
|
|
133.54
|
2.74e-02
|
GO:0048596
|
embryonic camera-type eye morphogenesis
|
|
4.87
|
2.94e-02
|
GO:0035556
|
intracellular signal transduction
|
|
7.28
|
3.10e-02
|
GO:0042325
|
regulation of phosphorylation
|
|
4.13
|
3.33e-02
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
|
7.07
|
3.63e-02
|
GO:0019220
|
regulation of phosphate metabolic process
|
|
7.07
|
3.63e-02
|
GO:0051174
|
regulation of phosphorus metabolic process
|
|
2.94
|
4.02e-02
|
GO:0007165
|
signal transduction
|
|
3.97
|
4.55e-02
|
GO:0048583
|
regulation of response to stimulus
|
|
13.90
|
4.63e-02
|
GO:0071375
|
cellular response to peptide hormone stimulus
|
|
13.70
|
4.89e-02
|
GO:0032868
|
response to insulin stimulus
|
|
4.52
|
4.98e-02
|
GO:0065009
|
regulation of molecular function
|