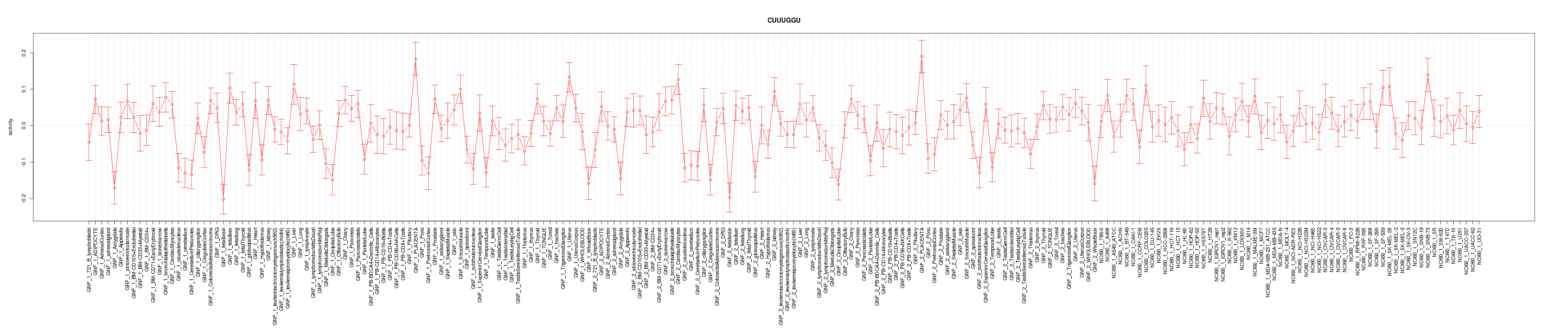
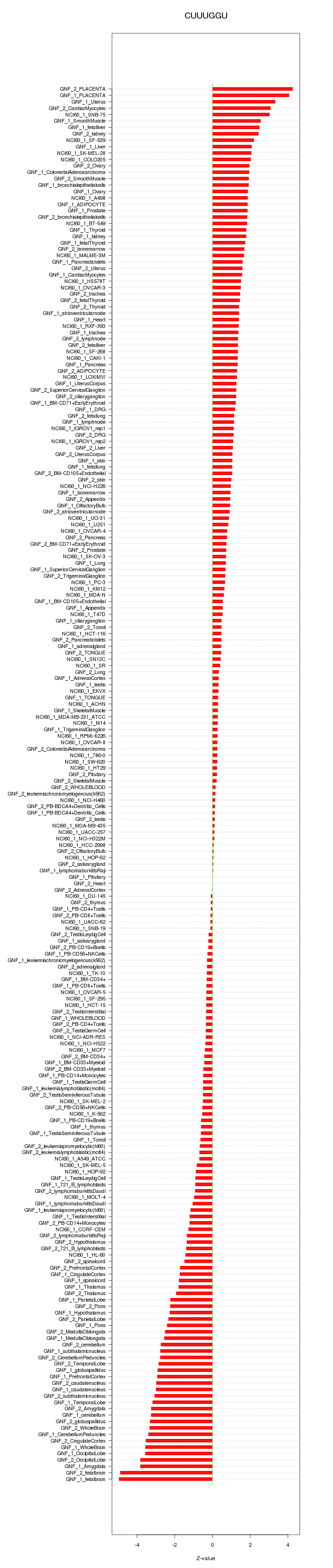
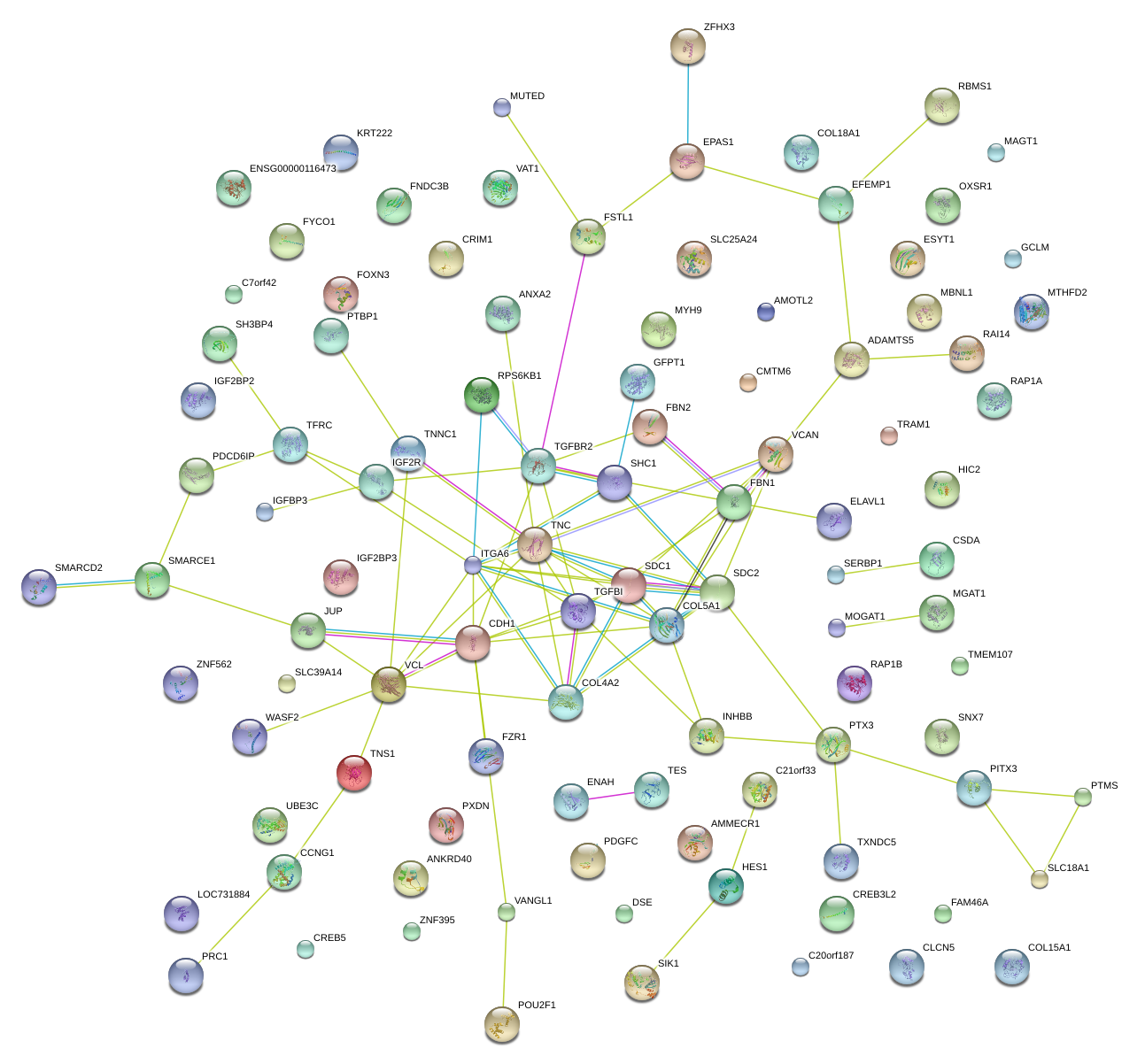
|
chr7_-_45960738
|
26.922
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr3_-_120169493
|
22.229
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr8_+_22224997
|
21.016
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr13_+_110959592
|
20.945
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr12_-_10875848
|
20.811
|
NM_001145426
NM_003651
|
CSDA
|
cold shock domain protein A
|
|
chr5_+_34687663
|
20.065
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr7_-_137686814
|
19.012
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr3_-_32544358
|
18.640
|
NM_017801
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr15_-_60690115
|
18.626
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr2_-_20424627
|
18.587
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr3_-_185542807
|
18.252
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr2_-_20425166
|
17.834
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr5_+_34684611
|
17.652
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr2_+_74425689
|
17.374
|
NM_006636
|
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase
|
|
chr2_+_235860616
|
16.966
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr6_-_7910716
|
16.806
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr2_-_161350304
|
16.704
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr3_+_171757413
|
16.684
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_-_154943001
|
16.253
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr21_-_44846927
|
15.725
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr6_-_7910278
|
15.533
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr5_+_135364583
|
15.418
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr5_+_34656367
|
14.954
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr19_-_8070468
|
14.770
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr15_-_48937795
|
14.683
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr6_-_82462374
|
14.531
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr21_+_46825001
|
14.492
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr5_+_34656595
|
14.400
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr9_-_117880411
|
14.088
|
NM_002160
|
TNC
|
tenascin C
|
|
chr3_-_195808958
|
14.014
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chrX_-_109683457
|
13.897
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr3_-_195808997
|
13.705
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_+_167298280
|
13.510
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_+_797390
|
13.433
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr2_-_1748285
|
13.214
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr4_-_157892545
|
13.080
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_-_69614321
|
13.041
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr3_+_171758289
|
12.981
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr8_+_22224758
|
12.930
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr8_-_71520620
|
12.677
|
NM_014294
|
TRAM1
|
translocation associated membrane protein 1
|
|
chr1_-_154946859
|
12.647
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chrX_-_109561314
|
12.261
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr9_+_137533643
|
11.857
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr5_+_162864571
|
11.756
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr2_-_56151273
|
11.650
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr21_+_46875402
|
11.475
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr9_+_101705710
|
11.336
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr17_-_39942937
|
11.281
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr1_-_108735362
|
11.255
|
NM_213651
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24
|
|
chr16_+_68771192
|
10.690
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr2_+_46524448
|
10.627
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chrX_-_77150907
|
10.626
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr10_+_75757847
|
10.521
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr8_-_28243941
|
10.423
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr1_-_108742963
|
10.277
|
NM_013386
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24
|
|
chr12_+_69004614
|
10.245
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr7_+_28725597
|
10.234
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_-_127873734
|
10.142
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr22_-_36784055
|
10.004
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr14_-_89883306
|
9.826
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr2_+_121103670
|
9.765
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr6_+_160390128
|
9.743
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr2_-_218808705
|
9.611
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr7_+_66386129
|
9.191
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr1_-_225840660
|
9.154
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr3_-_134093235
|
9.049
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr3_+_151985828
|
9.029
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr3_+_157154579
|
8.974
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr17_-_61920328
|
8.930
|
NM_001098426
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr16_-_73092533
|
8.901
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr7_+_115850546
|
8.898
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr3_+_152017193
|
8.830
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr3_+_193853930
|
8.750
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_+_30647901
|
8.747
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr1_+_167189948
|
8.701
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr15_-_91537724
|
8.651
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr6_+_116601282
|
8.636
|
NM_001080976
|
DSE
|
dermatan sulfate epimerase
|
|
chrX_+_49832214
|
8.623
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_-_27816676
|
8.585
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr2_+_36583369
|
8.464
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr12_+_56521953
|
8.455
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr17_-_48785016
|
8.404
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr7_+_28452143
|
8.401
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_-_94374986
|
8.271
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr17_-_41174431
|
8.194
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr3_+_33840060
|
8.151
|
NM_001162429
NM_013374
|
PDCD6IP
|
programmed cell death 6 interacting protein
|
|
chr17_-_38821286
|
8.147
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr2_+_173292306
|
8.145
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr3_+_38206968
|
8.130
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr5_-_180230889
|
8.129
|
NM_001114619
NM_001114620
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr21_-_28339405
|
8.068
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr7_+_28475233
|
7.984
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_-_67896098
|
7.973
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr5_+_82767492
|
7.967
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr6_+_116692098
|
7.904
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr7_+_156931631
|
7.844
|
NM_014671
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr22_+_21771661
|
7.840
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr17_-_38804037
|
7.753
|
NM_003079
|
SMARCE1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1
|
|
chr8_+_97505876
|
7.655
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr3_-_46037299
|
7.649
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr1_+_99127146
|
7.634
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr7_-_23509972
|
7.552
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr7_+_115863004
|
7.499
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr1_+_116184573
|
7.456
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr15_-_101792125
|
7.431
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr6_+_136172802
|
7.424
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr5_+_138089073
|
7.424
|
NM_001903
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa
|
|
chrX_-_153718984
|
7.372
|
NM_001142391
NM_001142392
NM_019848
|
SLC10A3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3
|
|
chr16_+_69220940
|
7.292
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr9_-_16870719
|
7.288
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr1_+_16693582
|
7.220
|
NM_001114600
NM_015609
|
C1orf144
|
chromosome 1 open reading frame 144
|
|
chr7_-_139876718
|
7.043
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr5_+_118691549
|
6.966
|
NM_014350
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr7_-_129592711
|
6.932
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr12_+_51157636
|
6.870
|
NM_005171
|
ATF1
|
activating transcription factor 1
|
|
chr17_+_37026111
|
6.847
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr2_-_110371669
|
6.777
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr13_+_113863054
|
6.745
|
NM_003589
|
CUL4A
|
cullin 4A
|
|
chr11_-_8680382
|
6.732
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr1_+_26737232
|
6.725
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr10_-_14372807
|
6.701
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_-_108507474
|
6.692
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_+_155100304
|
6.555
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr3_-_107809754
|
6.554
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr1_+_193091057
|
6.473
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chrX_+_49687212
|
6.467
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr12_-_54653317
|
6.466
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr13_+_73633097
|
6.440
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr13_+_96329392
|
6.417
|
NM_006260
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr2_+_178257371
|
6.403
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr15_+_40733302
|
6.299
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chrX_+_12809473
|
6.295
|
NM_001039091
NM_002765
|
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2
|
|
chr17_-_1395950
|
6.285
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr19_+_16222434
|
6.262
|
NM_005370
|
RAB8A
|
RAB8A, member RAS oncogene family
|
|
chr2_+_48541767
|
6.208
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr1_-_108231122
|
6.206
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_+_154797435
|
6.175
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_116185249
|
6.169
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr5_-_180230047
|
6.156
|
NM_002406
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr14_-_34931413
|
6.138
|
NM_138288
|
SPTSSA
|
serine palmitoyltransferase, small subunit A
|
|
chr4_+_57774035
|
6.122
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr7_+_101460881
|
6.069
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr1_+_65613211
|
6.050
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_+_113162074
|
5.999
|
NM_006135
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1
|
|
chr5_-_180237114
|
5.982
|
NM_001114617
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr11_+_101980736
|
5.920
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr5_+_118604417
|
5.900
|
NM_001077654
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr14_+_103058988
|
5.866
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr3_-_87040246
|
5.853
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr13_-_33780142
|
5.754
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_+_1728256
|
5.749
|
NM_144570
|
HN1L
|
hematological and neurological expressed 1-like
|
|
chr7_-_129592794
|
5.728
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr18_+_33234532
|
5.662
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr1_+_145438437
|
5.657
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr5_-_131562934
|
5.640
|
NM_001017973
NM_004199
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr5_-_133968463
|
5.604
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr9_+_100745456
|
5.604
|
NM_006401
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr12_-_102872329
|
5.599
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_-_32174397
|
5.591
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr9_+_137218313
|
5.582
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr3_-_11623829
|
5.558
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_7831326
|
5.511
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr2_+_75059781
|
5.507
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr9_-_107690526
|
5.504
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr11_+_60681370
|
5.495
|
NM_024092
|
TMEM109
|
transmembrane protein 109
|
|
chr12_-_89746244
|
5.486
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr10_+_69644938
|
5.443
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr7_+_28338939
|
5.406
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_58240746
|
5.399
|
NM_005730
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr5_+_149340287
|
5.357
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr13_+_113863926
|
5.349
|
NM_001008895
|
CUL4A
|
cullin 4A
|
|
chr17_-_31203889
|
5.322
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr11_-_46142955
|
5.309
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr20_+_51588945
|
5.308
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_+_25071759
|
5.294
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr3_+_154797644
|
5.265
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr15_-_78369825
|
5.257
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr8_+_37654395
|
5.187
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr4_+_17812524
|
5.151
|
NM_022346
|
NCAPG
|
non-SMC condensin I complex, subunit G
|
|
chr2_-_37544165
|
5.140
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr5_-_83016850
|
5.139
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr15_-_60884615
|
5.135
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr4_+_41937105
|
5.089
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr5_-_131563481
|
5.077
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr16_-_85045119
|
5.076
|
NM_001145548
NM_017740
|
ZDHHC7
|
zinc finger, DHHC-type containing 7
|
|
chr1_+_155108276
|
5.028
|
NM_001122837
NM_001122839
NM_018845
|
SLC50A1
|
solute carrier family 50 (sugar transporter), member 1
|
|
chr17_-_1394849
|
5.016
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr13_-_33859835
|
5.003
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr15_+_91411884
|
5.001
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr3_+_154797910
|
4.977
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_65613771
|
4.964
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr11_+_101983170
|
4.956
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr5_-_180236816
|
4.889
|
NM_001114618
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr16_-_73082169
|
4.879
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr1_+_151512760
|
4.872
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|
|
chr3_+_57994087
|
4.870
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr10_-_61666236
|
4.842
|
NM_005436
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr15_-_56285732
|
4.840
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr3_+_180630054
|
4.834
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr1_-_169455099
|
4.831
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|