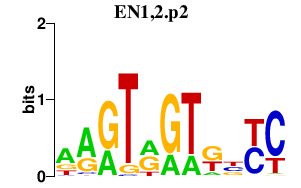
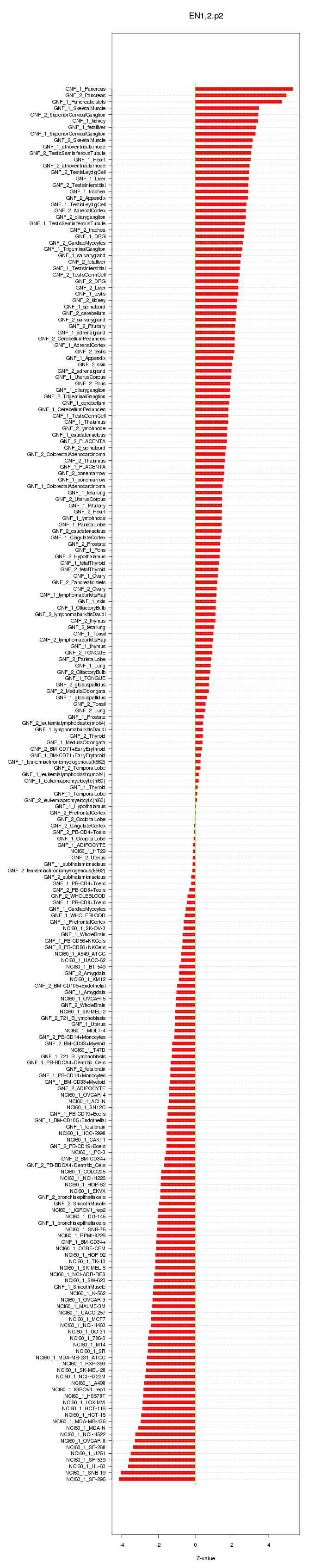
|
chr9_+_139872014
|
54.739
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr9_+_139871954
|
52.893
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr11_-_5248180
|
52.413
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr14_-_21271002
|
46.105
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr14_-_21270537
|
37.932
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr12_+_21679190
|
27.923
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr8_-_87755880
|
27.037
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr5_+_40909558
|
25.191
|
NM_000587
|
C7
|
complement component 7
|
|
chr3_-_38835500
|
24.054
|
NM_006514
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit
|
|
chr20_+_34556528
|
22.600
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr1_-_163172624
|
22.187
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr2_+_103035148
|
22.135
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr14_-_53620032
|
21.985
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr5_+_40909617
|
21.924
|
|
C7
|
complement component 7
|
|
chr16_+_31085751
|
21.876
|
|
ZNF646
|
zinc finger protein 646
|
|
chr6_-_32013806
|
20.334
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr17_+_4046417
|
20.074
|
NM_144611
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr4_+_74281884
|
19.996
|
|
ALB
|
albumin
|
|
chr15_-_74659926
|
19.902
|
NM_000781
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr16_+_31085722
|
19.889
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr12_-_105322056
|
19.428
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr1_-_163113129
|
19.394
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr7_+_117864711
|
19.247
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr11_-_35547144
|
18.607
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr17_+_45810609
|
18.548
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr10_+_17851342
|
18.332
|
NM_002438
|
MRC1
|
mannose receptor, C type 1
|
|
chr11_+_49053151
|
18.317
|
NM_001206626
|
LOC283116
|
tripartite motif-containing protein LOC642612-like
|
|
chr12_-_11548485
|
17.859
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr17_-_4046298
|
17.770
|
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1
|
|
chr4_-_39033981
|
17.647
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr10_+_75532252
|
17.071
|
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr10_+_105037441
|
16.899
|
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr1_-_212208779
|
16.855
|
NM_001199809
NM_001199811
NM_001199812
NM_015434
|
INTS7
|
integrator complex subunit 7
|
|
chr16_-_31085503
|
16.660
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr1_+_160765927
|
16.364
|
NM_001033667
NM_002348
|
LY9
|
lymphocyte antigen 9
|
|
chr6_+_41303527
|
16.217
|
NM_001199509
NM_001199510
NM_004828
|
NCR2
|
natural cytotoxicity triggering receptor 2
|
|
chr17_-_29624249
|
16.133
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr17_-_4046190
|
16.091
|
|
|
|
|
chr14_-_93799371
|
15.374
|
NM_001002860
NM_018167
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr1_+_86934525
|
15.346
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr16_-_20364029
|
15.326
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr1_+_171154387
|
15.194
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr1_-_155145732
|
15.179
|
NM_173852
|
KRTCAP2
|
keratinocyte associated protein 2
|
|
chr9_-_34691135
|
15.086
|
NM_006274
|
CCL19
|
chemokine (C-C motif) ligand 19
|
|
chr10_-_104179468
|
14.877
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr11_+_5009420
|
14.505
|
NM_021801
|
MMP26
|
matrix metallopeptidase 26
|
|
chrX_+_128913897
|
14.473
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr6_+_32407618
|
14.287
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr19_-_46295785
|
14.055
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr2_-_217540722
|
13.680
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr6_-_52668599
|
13.571
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chrX_+_103031433
|
13.371
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_38420690
|
13.345
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr2_-_192015706
|
13.095
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr2_-_163008913
|
13.016
|
NM_002054
|
GCG
|
glucagon
|
|
chr8_-_82395401
|
12.970
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chrX_+_128913881
|
12.836
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr17_-_43487747
|
12.790
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr16_-_3627353
|
12.746
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr8_+_11141983
|
12.698
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr8_+_11142314
|
12.628
|
|
MTMR9
|
myotubularin related protein 9
|
|
chr3_+_138066489
|
11.990
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr17_-_56591900
|
11.923
|
|
MTMR4
|
myotubularin related protein 4
|
|
chr2_-_152590983
|
11.876
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr22_+_23054862
|
11.836
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr11_-_57004674
|
11.711
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr17_+_37821598
|
11.690
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chrX_+_103031797
|
11.663
|
|
PLP1
|
proteolipid protein 1
|
|
chr7_-_44105162
|
11.401
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr1_+_32050510
|
11.370
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr1_-_152086555
|
11.343
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr12_-_13103317
|
11.342
|
NM_018654
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D
|
|
chr12_-_6486522
|
11.311
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr6_+_31913852
|
11.249
|
|
CFB
|
complement factor B
|
|
chr19_+_17326196
|
11.223
|
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr15_+_93443418
|
11.206
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_+_124317884
|
11.126
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr12_-_91398744
|
11.061
|
|
EPYC
|
epiphycan
|
|
chr2_+_60983354
|
10.999
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr5_+_140474198
|
10.997
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr3_-_187452669
|
10.958
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chrX_+_152990322
|
10.923
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr1_+_15802595
|
10.880
|
NM_015849
|
CELA2B
|
chymotrypsin-like elastase family, member 2B
|
|
chrX_-_65859107
|
10.874
|
NM_001199687
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chrX_+_103031753
|
10.844
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_110366304
|
10.684
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr8_-_33424642
|
10.670
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr2_+_79347610
|
10.546
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr12_-_57630867
|
10.531
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr8_-_57358511
|
10.527
|
NM_006211
|
PENK
|
proenkephalin
|
|
chr9_-_34729463
|
10.475
|
NM_001141917
|
FAM205A
|
family with sequence similarity 205, member A
|
|
chr15_-_65412175
|
10.468
|
|
PDCD7
|
programmed cell death 7
|
|
chr8_+_133879202
|
10.410
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr19_+_41509825
|
10.329
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr6_+_101846668
|
10.306
|
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr10_+_118305427
|
10.268
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr1_-_150587879
|
10.256
|
|
|
|
|
chr1_-_163113938
|
10.237
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chrX_+_18709044
|
10.207
|
NM_006240
NM_152224
NM_152226
|
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1
|
|
chr5_+_54455945
|
10.176
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr3_-_46505077
|
10.161
|
NM_001199149
|
LTF
|
lactotransferrin
|
|
chr19_-_48823331
|
9.973
|
NM_144577
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr4_-_76928601
|
9.866
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chrX_-_7895424
|
9.839
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr14_+_95078713
|
9.780
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr4_+_71337833
|
9.762
|
NM_152291
|
MUC7
|
mucin 7, secreted
|
|
chr8_+_22210311
|
9.761
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr5_-_141993691
|
9.755
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_9834201
|
9.741
|
NM_001198793
NM_005718
|
ARPC4-TTLL3
ARPC4
|
ARPC4-TTLL3 readthrough
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr6_+_106546736
|
9.741
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr10_-_104597079
|
9.680
|
NM_000102
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1
|
|
chr20_-_1472004
|
9.571
|
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr5_+_68800003
|
9.571
|
NM_001205255
|
OCLN
|
occludin
|
|
chr6_-_32977312
|
9.543
|
|
HLA-DOA
|
major histocompatibility complex, class II, DO alpha
|
|
chr8_+_40010986
|
9.542
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr1_+_32043745
|
9.491
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr1_-_203320249
|
9.368
|
|
|
|
|
chr9_-_123812535
|
9.344
|
NM_001735
|
C5
|
complement component 5
|
|
chr19_-_54327428
|
9.340
|
NM_144687
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr2_+_233390869
|
9.332
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr9_+_105757592
|
9.317
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr4_+_40198526
|
9.200
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr16_+_67381252
|
9.175
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr4_-_17806820
|
9.138
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr6_-_52628272
|
9.130
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr4_+_72102249
|
8.999
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_-_27701302
|
8.946
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chr9_-_79520878
|
8.887
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr4_+_78432905
|
8.806
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr3_+_183903862
|
8.780
|
NM_018358
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr12_+_56732658
|
8.763
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr12_-_102591547
|
8.730
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr4_+_71337674
|
8.728
|
NM_001145007
|
MUC7
|
mucin 7, secreted
|
|
chr22_+_22599199
|
8.679
|
NM_007128
|
VPREB1
|
pre-B lymphocyte 1
|
|
chr1_-_216978708
|
8.596
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_+_128427856
|
8.464
|
NM_001159542
|
POU5F1B
|
POU class 5 homeobox 1B
|
|
chr17_-_64225496
|
8.331
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr11_-_5255712
|
8.292
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chrX_+_50653734
|
8.266
|
NM_005448
|
BMP15
|
bone morphogenetic protein 15
|
|
chr21_-_40685418
|
8.245
|
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr19_-_41356193
|
8.244
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr15_+_68908878
|
8.225
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr5_+_180415844
|
8.222
|
NM_197975
|
BTNL3
|
butyrophilin-like 3
|
|
chr10_+_118380451
|
8.214
|
NM_005396
|
PNLIPRP2
|
pancreatic lipase-related protein 2
|
|
chr4_+_74269971
|
8.199
|
NM_000477
|
ALB
|
albumin
|
|
chr10_-_62149487
|
8.157
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_119470336
|
8.155
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr5_+_131993864
|
8.149
|
NM_002188
|
IL13
|
interleukin 13
|
|
chr21_+_34697277
|
8.128
|
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1
|
|
chr5_+_55062712
|
8.128
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr6_-_31939782
|
8.078
|
NM_005510
|
DOM3Z
|
dom-3 homolog Z (C. elegans)
|
|
chr8_-_27115935
|
8.065
|
|
STMN4
|
stathmin-like 4
|
|
chr10_+_49609641
|
8.060
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr6_+_31583766
|
8.052
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr2_+_219246751
|
8.049
|
NM_000578
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr17_+_61562177
|
7.960
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr6_+_31913708
|
7.941
|
NM_001710
|
CFB
|
complement factor B
|
|
chr22_+_40297085
|
7.923
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr22_+_22712091
|
7.910
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr9_-_21077804
|
7.902
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr12_-_11508509
|
7.873
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chrX_-_104465349
|
7.865
|
NM_031274
|
TEX13A
|
testis expressed 13A
|
|
chr6_+_42123143
|
7.853
|
NM_000409
|
GUCA1A
|
guanylate cyclase activator 1A (retina)
|
|
chr7_+_107110501
|
7.801
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr18_-_13915534
|
7.794
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr3_-_9834357
|
7.741
|
NM_006354
NM_133480
|
TADA3
|
transcriptional adaptor 3
|
|
chr13_-_73356058
|
7.734
|
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr11_-_84028379
|
7.729
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_+_16348365
|
7.728
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr6_+_29364331
|
7.686
|
NM_013936
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2
|
|
chr1_-_151798552
|
7.651
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr17_-_39093671
|
7.645
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr22_+_40297183
|
7.574
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr10_+_105036783
|
7.568
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chrX_+_49593905
|
7.507
|
NM_007003
|
PAGE4
|
P antigen family, member 4 (prostate associated)
|
|
chr6_+_161123224
|
7.497
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr1_-_161279724
|
7.473
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr5_-_64777703
|
7.467
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr2_+_230787195
|
7.463
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr20_+_47895168
|
7.454
|
|
ZNFX1-AS1
|
ZNFX1 antisense RNA 1 (non-protein coding)
|
|
chr20_-_47894570
|
7.370
|
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr7_+_129906702
|
7.345
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr19_+_35739778
|
7.312
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr13_+_42712177
|
7.285
|
NM_001204505
NM_001204506
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr14_+_95047730
|
7.284
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr1_-_12908141
|
7.271
|
NM_001146181
|
HNRNPCL1
LOC649330
|
heterogeneous nuclear ribonucleoprotein C-like 1
heterogeneous nuclear ribonucleoprotein C-like
|
|
chr12_-_120765555
|
7.247
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr2_+_33738941
|
7.211
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr14_-_80677839
|
7.171
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr5_-_24645084
|
7.153
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr19_+_41594367
|
7.151
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr10_-_7682741
|
7.136
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr21_+_34697197
|
7.103
|
NM_000629
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1
|
|
chr7_+_128578246
|
7.100
|
NM_032643
|
IRF5
|
interferon regulatory factor 5
|
|
chr20_-_1472168
|
7.099
|
NM_001122962
NM_001134836
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr19_+_15852200
|
7.082
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr19_-_58067724
|
7.066
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr3_+_52813936
|
7.054
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr21_-_40685697
|
7.049
|
NM_001007246
NM_018963
NM_033656
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr6_+_33043702
|
7.047
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|