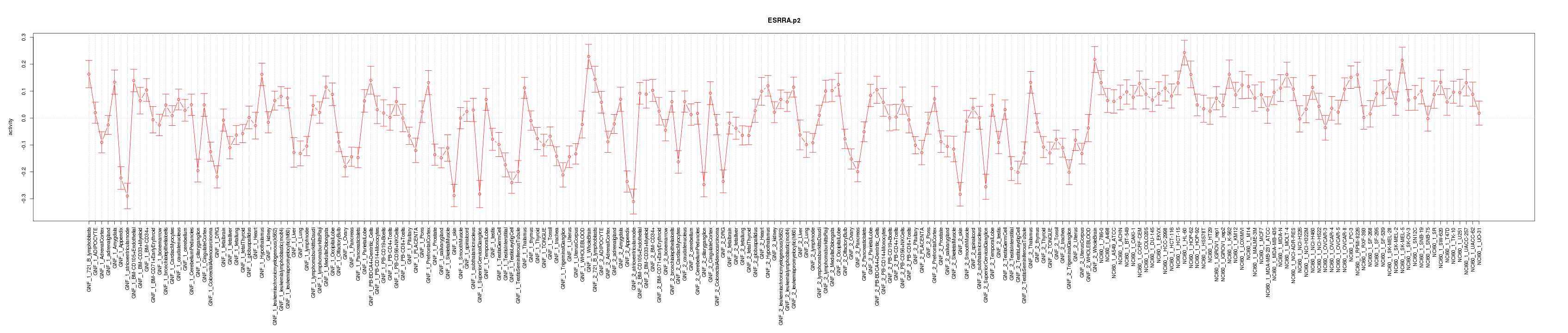
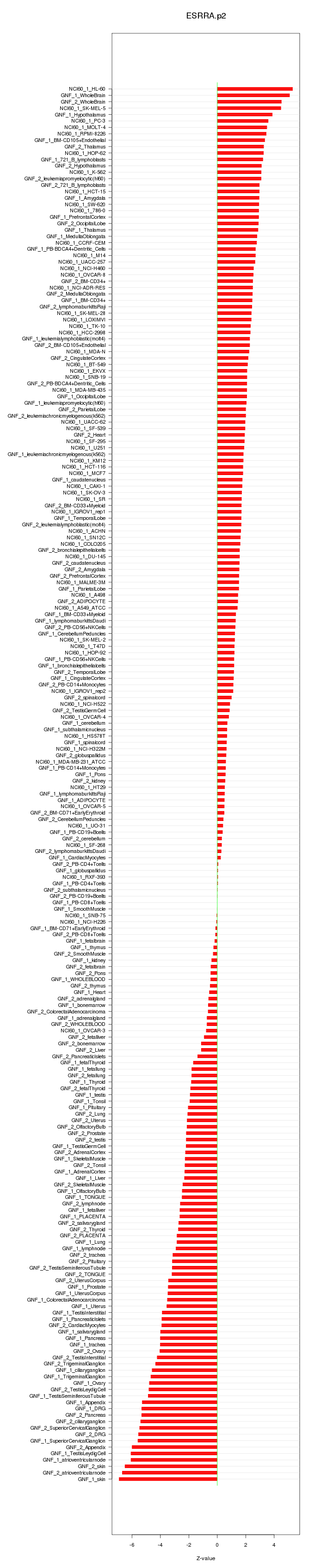
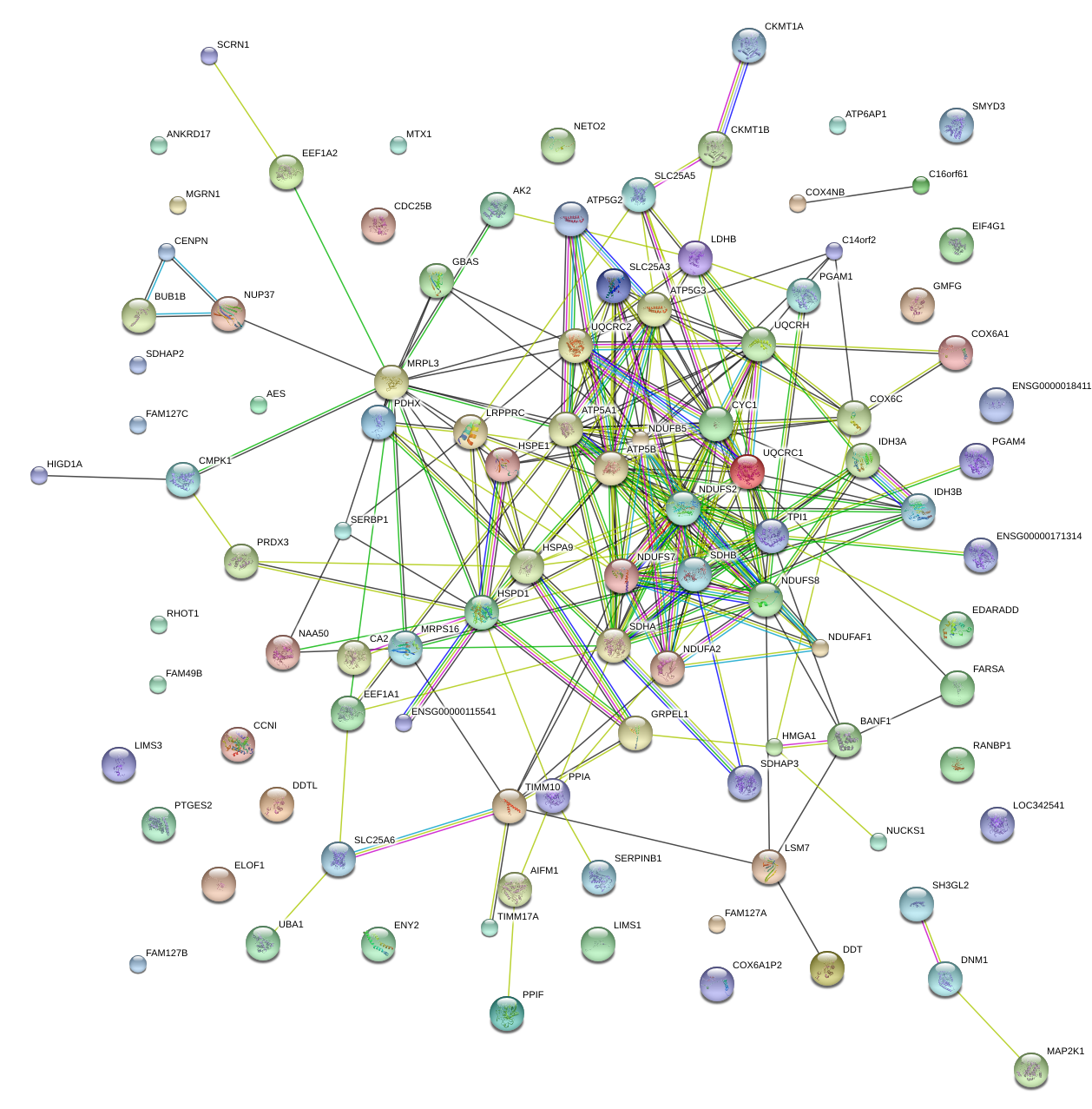
|
chr8_+_145149985
|
82.272
|
|
CYC1
|
cytochrome c-1
|
|
chr2_-_176046120
|
81.835
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr10_+_81107259
|
80.838
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_+_81107242
|
80.779
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr5_+_218428
|
50.854
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chrX_+_118602391
|
47.372
|
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr5_+_218441
|
45.038
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr8_+_145149936
|
42.879
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr3_-_131221649
|
42.847
|
NM_007208
|
MRPL3
|
mitochondrial ribosomal protein L3
|
|
chr8_+_145149978
|
41.774
|
|
CYC1
|
cytochrome c-1
|
|
chr10_+_81107219
|
40.200
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_+_81107228
|
40.019
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr12_-_21810720
|
37.995
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr2_-_44223131
|
37.915
|
NM_133259
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr12_-_21810779
|
37.865
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810733
|
37.493
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810754
|
37.309
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr19_-_13044459
|
36.909
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr16_+_21964700
|
36.800
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr20_+_3776905
|
36.716
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr12_-_21810769
|
36.692
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr16_+_21964677
|
36.637
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr20_+_3776385
|
36.278
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr10_-_120938276
|
35.594
|
|
PRDX3
|
peroxiredoxin 3
|
|
chr16_+_21964608
|
35.355
|
NM_003366
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr7_-_30029904
|
35.022
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr12_+_6976694
|
35.021
|
|
TPI1
|
triosephosphate isomerase 1
|
|
chr18_-_43678233
|
34.996
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr12_+_6976583
|
33.863
|
NM_001159287
NM_000365
|
TPI1
|
triosephosphate isomerase 1
|
|
chr1_+_46769336
|
33.770
|
NM_006004
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein
|
|
chr2_-_44223112
|
32.950
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr15_+_40453263
|
32.041
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr14_-_104387837
|
32.023
|
|
C14orf2
|
chromosome 14 open reading frame 2
|
|
chr10_+_99185977
|
31.991
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr5_+_218355
|
31.846
|
NM_004168
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr8_+_110346549
|
31.746
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr11_+_65769886
|
30.606
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr8_-_131028674
|
30.477
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr10_-_120938337
|
30.397
|
NM_006793
NM_014098
|
PRDX3
|
peroxiredoxin 3
|
|
chr11_+_65769956
|
30.276
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr5_+_218447
|
30.134
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr9_-_130889993
|
29.914
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chrX_-_134156488
|
29.859
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr2_+_198365012
|
29.809
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr16_-_81040439
|
29.718
|
NM_020188
|
C16orf61
|
chromosome 16 open reading frame 61
|
|
chr3_+_179322572
|
29.661
|
NM_001199957
NM_001199958
NM_002492
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
|
|
chr12_+_120875891
|
29.309
|
NM_004373
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1
|
|
chr3_-_42845911
|
29.290
|
NM_001099668
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr2_-_44223039
|
29.226
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr7_-_30029284
|
29.106
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr22_+_20105145
|
28.847
|
|
RANBP1
|
RAN binding protein 1
|
|
chr8_-_100905910
|
28.618
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr11_-_57298180
|
28.593
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr4_-_74088773
|
28.442
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr12_-_54070108
|
28.403
|
NM_005176
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9)
|
|
chr5_-_140027167
|
27.829
|
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa
|
|
chr3_-_48647085
|
27.795
|
NM_003365
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I
|
|
chr1_-_17380486
|
27.456
|
NM_003000
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr3_-_42845946
|
27.231
|
NM_001099669
NM_014056
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr1_-_205719340
|
27.108
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr18_-_43678195
|
26.987
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr19_-_39826676
|
26.946
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_48647052
|
26.869
|
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I
|
|
chr16_+_4674829
|
26.796
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr16_+_4674819
|
26.749
|
NM_001142289
NM_001142290
NM_001142291
NM_015246
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr1_+_161172174
|
26.725
|
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr1_+_161172163
|
26.704
|
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr2_+_198365113
|
26.638
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr10_+_99186012
|
26.408
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr15_+_78441733
|
26.235
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr10_-_75012241
|
26.186
|
NM_016065
|
MRPS16
|
mitochondrial ribosomal protein S16
|
|
chr18_-_43678230
|
25.722
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr19_-_2328566
|
25.716
|
NM_016199
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_+_47799447
|
25.648
|
NM_001136140
NM_016308
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr11_+_34938112
|
25.506
|
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr9_+_130965666
|
25.254
|
|
DNM1
|
dynamin 1
|
|
chr18_-_43678251
|
24.860
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr1_-_246580710
|
24.827
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chr16_-_85832900
|
24.775
|
NM_001142288
NM_006067
|
COX4NB
|
COX4 neighbor
|
|
chr6_+_34206567
|
24.754
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr20_-_2641476
|
24.565
|
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr1_+_236558715
|
24.300
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr2_+_109237679
|
24.300
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_-_131028833
|
24.299
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr8_-_100905865
|
24.265
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr12_-_102512277
|
24.113
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr22_+_20105095
|
24.032
|
|
RANBP1
|
RAN binding protein 1
|
|
chr2_-_198364522
|
24.021
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr19_-_3062911
|
24.000
|
NM_198969
|
AES
|
amino-terminal enhancer of split
|
|
chr5_-_137911039
|
23.987
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr12_-_57039785
|
23.982
|
NM_001686
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr22_+_20105076
|
23.815
|
|
RANBP1
|
RAN binding protein 1
|
|
chrX_+_47053192
|
23.739
|
NM_003334
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr12_+_98987674
|
23.642
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chrX_-_129299680
|
23.488
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr7_+_44836273
|
23.357
|
|
PPIA
|
peptidylprolyl isomerase A (cyclophilin A)
|
|
chr22_-_24316602
|
23.245
|
NM_001084392
|
DDT
|
D-dopachrome tautomerase
|
|
chr22_+_24309025
|
23.219
|
NM_001084393
|
DDTL
|
D-dopachrome tautomerase-like
|
|
chr4_-_7069755
|
23.188
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr10_+_99186040
|
23.184
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr10_+_99186042
|
23.049
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr20_-_62130390
|
22.940
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr5_-_137911005
|
22.750
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr19_+_1383898
|
22.605
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chrX_+_47053227
|
22.487
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr2_-_198364560
|
22.275
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr6_-_2840683
|
22.056
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr16_-_2723374
|
22.053
|
|
ERVK13-1
|
endogenous retrovirus group K13, member 1
|
|
chrX_+_47053213
|
22.019
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr12_-_54069856
|
21.951
|
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9)
|
|
chr19_+_1383904
|
21.899
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chrX_-_129299629
|
21.831
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr1_+_47799488
|
21.791
|
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr15_-_41694619
|
21.619
|
NM_016013
|
NDUFAF1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1
|
|
chrX_+_47053271
|
21.452
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr2_-_198364623
|
21.390
|
NM_002156
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr9_+_17578952
|
21.147
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr1_+_201924618
|
20.904
|
NM_006335
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast)
|
|
chr1_-_33502470
|
20.855
|
|
AK2
|
adenylate kinase 2
|
|
chr12_-_57039736
|
20.575
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr3_+_195384941
|
20.534
|
|
SDHAP2
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2
|
|
chrX_-_129299803
|
20.486
|
NM_001130847
NM_004208
NM_145812
NM_145813
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr1_-_33502452
|
20.453
|
|
AK2
|
adenylate kinase 2
|
|
chr10_+_99186017
|
20.399
|
NM_002629
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chrX_+_47053302
|
20.369
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr7_+_56032065
|
20.327
|
NM_001202469
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chr19_+_1383877
|
20.322
|
NM_024407
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chrX_+_47053234
|
20.277
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr19_-_13044528
|
20.260
|
NM_004461
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr16_-_47177902
|
20.209
|
NM_001201477
NM_018092
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr15_+_66694216
|
20.114
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr16_+_81040515
|
20.018
|
|
CENPN
|
centromere protein N
|
|
chr9_-_130889965
|
20.001
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr11_+_67798077
|
19.935
|
NM_002496
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
|
|
chrX_+_153657119
|
19.656
|
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1
|
|
chr19_-_3062191
|
19.424
|
NM_001130
NM_198970
|
AES
|
amino-terminal enhancer of split
|
|
chr16_-_47177832
|
19.388
|
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr1_+_155179005
|
19.373
|
|
MTX1
|
metaxin 1
|
|
chr22_-_43036606
|
19.279
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr1_-_33502464
|
19.187
|
|
AK2
|
adenylate kinase 2
|
|
chr17_+_30469567
|
19.031
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr3_-_113465101
|
18.927
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr8_+_86376130
|
18.788
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr17_+_30469541
|
18.750
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr3_+_184038050
|
18.709
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr8_-_131028646
|
18.520
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_-_67896098
|
18.476
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr10_+_99186004
|
18.318
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr1_+_161172125
|
18.302
|
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr15_+_43885251
|
18.167
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr16_-_28222700
|
17.961
|
|
XPO6
|
exportin 6
|
|
chr1_-_33502428
|
17.960
|
|
AK2
|
adenylate kinase 2
|
|
chr13_-_24007822
|
17.906
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr11_+_73498913
|
17.802
|
NM_016055
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr22_-_47134130
|
17.769
|
NM_022766
|
CERK
|
ceramide kinase
|
|
chr22_-_36681757
|
17.744
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr10_+_62538088
|
17.712
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr20_+_57466248
|
17.661
|
NM_000516
NM_001077488
NM_001077489
NM_080426
|
GNAS
|
GNAS complex locus
|
|
chr20_-_31989303
|
17.631
|
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr6_-_97345760
|
17.503
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr7_+_99686609
|
17.431
|
|
COPS6
|
COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis)
|
|
chr14_+_57735605
|
17.320
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr17_+_79670495
|
17.207
|
|
MRPL12
|
mitochondrial ribosomal protein L12
|
|
chr19_-_39826625
|
17.115
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_10028512
|
16.972
|
NM_018447
|
TMEM111
|
transmembrane protein 111
|
|
chr1_+_228270802
|
16.946
|
NM_001024227
NM_001024228
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr15_+_78441678
|
16.895
|
NM_005530
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr22_-_50524357
|
16.831
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr1_+_32757711
|
16.735
|
|
HDAC1
|
histone deacetylase 1
|
|
chr3_-_195808934
|
16.598
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr10_-_126107493
|
16.539
|
NM_000274
NM_001171814
|
OAT
|
ornithine aminotransferase
|
|
chr3_-_195808958
|
16.534
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr15_-_83735892
|
16.527
|
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr17_+_79670396
|
16.359
|
NM_002949
|
MRPL12
|
mitochondrial ribosomal protein L12
|
|
chr1_+_32757702
|
16.162
|
|
HDAC1
|
histone deacetylase 1
|
|
chr14_-_105261850
|
16.152
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr9_-_130890026
|
16.096
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr2_+_26467615
|
15.991
|
NM_000183
|
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit
|
|
chr18_-_43678273
|
15.968
|
NM_004046
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr17_+_46970128
|
15.963
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr19_+_54606141
|
15.896
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr9_-_130890007
|
15.838
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chrY_-_1460901
|
15.767
|
NM_001636
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6
|
|
chr5_+_85913762
|
15.563
|
NM_001867
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr10_-_118765087
|
15.495
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr14_+_100150596
|
15.486
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chrX_-_109683457
|
15.474
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr6_-_97345747
|
15.350
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr20_-_31989330
|
15.283
|
NM_016082
NM_016408
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr1_+_32757707
|
15.247
|
NM_004964
|
HDAC1
|
histone deacetylase 1
|
|
chr1_+_165600433
|
15.183
|
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr3_-_10028344
|
15.137
|
|
TMEM111
|
transmembrane protein 111
|
|
chr9_+_130965668
|
15.069
|
|
DNM1
|
dynamin 1
|
|
chr1_-_27732069
|
15.006
|
|
WASF2
|
WAS protein family, member 2
|
|
chr22_+_44351260
|
14.991
|
NM_015380
|
SAMM50
|
sorting and assembly machinery component 50 homolog (S. cerevisiae)
|
|
chr1_+_6094342
|
14.942
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr22_-_42486740
|
14.850
|
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa
|
|
chr1_-_67896081
|
14.691
|
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr19_+_8509802
|
14.676
|
NM_005968
NM_031203
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr11_+_67033889
|
14.636
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|