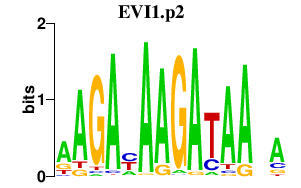
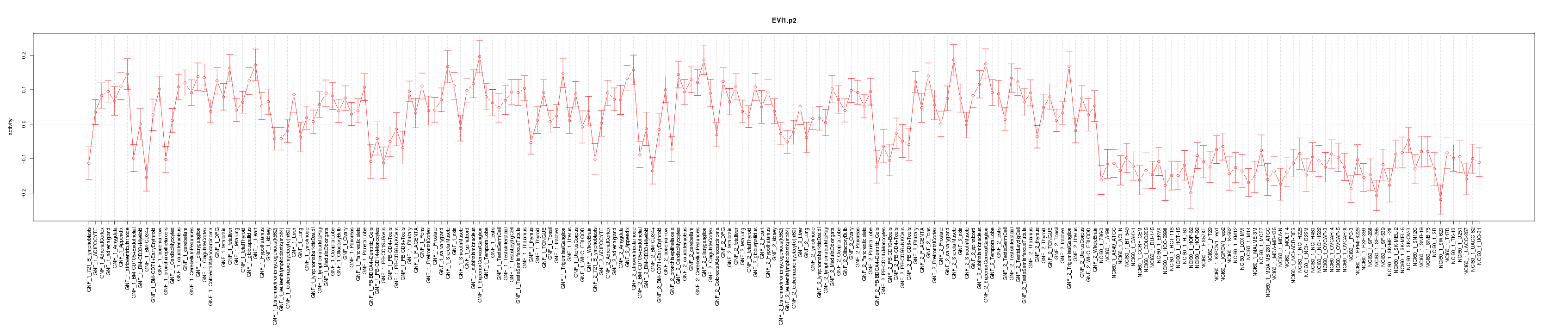
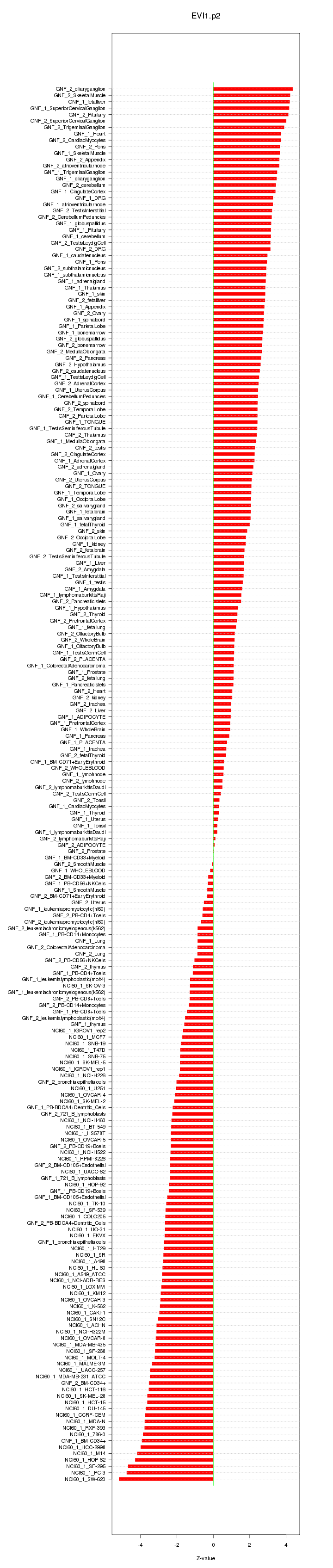
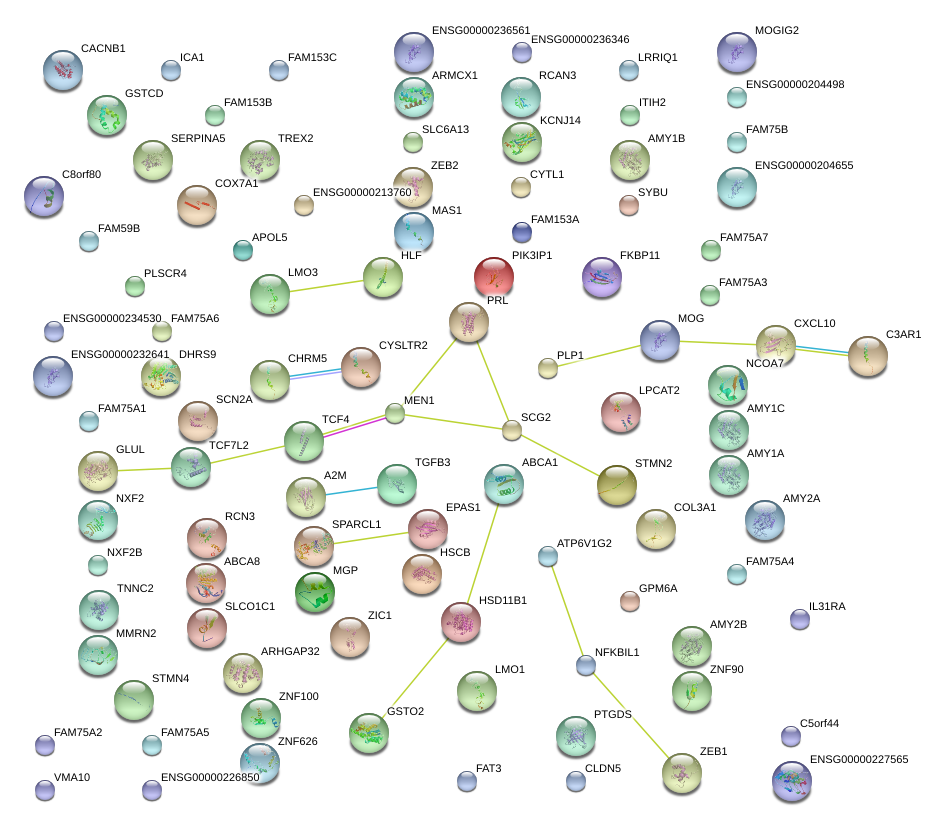
|
chr4_-_88450243
|
91.312
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr9_+_139871954
|
68.202
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr8_-_27115897
|
62.831
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr17_+_53344966
|
50.667
|
|
HLF
|
hepatic leukemia factor
|
|
chr2_-_224467095
|
49.880
|
|
SCG2
|
secretogranin II
|
|
chr12_-_15038724
|
48.480
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr6_-_31514620
|
43.243
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr8_-_27115935
|
41.642
|
|
STMN4
|
stathmin-like 4
|
|
chr10_+_106034886
|
39.622
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr14_-_76447445
|
39.253
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr19_-_36643770
|
38.626
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr2_-_224467049
|
38.550
|
|
SCG2
|
secretogranin II
|
|
chr1_-_93399531
|
37.088
|
|
|
|
|
chr10_+_7745341
|
36.641
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr15_+_66874506
|
35.756
|
|
|
|
|
chr10_+_7745325
|
30.903
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr2_-_224467142
|
30.842
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr17_-_66951381
|
30.409
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr12_-_16758312
|
29.744
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_+_31515391
|
28.886
|
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr8_-_27941387
|
28.637
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr2_-_89165652
|
28.571
|
|
|
|
|
chr2_-_145274916
|
28.421
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_-_16757944
|
28.195
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_80523376
|
27.948
|
|
STMN2
|
stathmin-like 2
|
|
chr5_+_140739589
|
27.204
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr6_-_22297729
|
27.162
|
NM_000948
|
PRL
|
prolactin
|
|
chr15_+_34261088
|
26.598
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr12_+_20848288
|
26.107
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr12_-_8218930
|
26.031
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr16_+_55542909
|
26.014
|
NM_017839
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2
|
|
chr12_-_8219001
|
25.900
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr10_+_7745284
|
25.774
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr11_+_92085261
|
25.523
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr10_-_88717319
|
25.318
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr22_+_29138018
|
25.276
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr6_-_116422408
|
25.274
|
|
|
|
|
chr6_+_31515352
|
25.269
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr4_+_106631966
|
24.912
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr8_-_110655418
|
24.319
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_189839132
|
23.777
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_371994
|
23.356
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chrX_+_101502093
|
23.222
|
NM_022053
NM_001099686
|
NXF2
NXF2B
|
nuclear RNA export factor 2
nuclear RNA export factor 2B
|
|
chr10_+_7745354
|
22.927
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr14_+_95047758
|
22.747
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chrX_+_103031753
|
22.695
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr12_-_9268440
|
22.571
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr5_+_55147206
|
22.450
|
NM_001242637
NM_001242638
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr5_+_175511848
|
21.957
|
NM_001079529
|
FAM153B
|
family with sequence similarity 153, member B
|
|
chrX_-_101694848
|
21.956
|
NM_022053
NM_001099686
|
NXF2
NXF2B
|
nuclear RNA export factor 2
nuclear RNA export factor 2B
|
|
chr19_-_20844312
|
21.405
|
NM_001076675
NM_145297
|
ZNF626
|
zinc finger protein 626
|
|
chr22_-_31688329
|
21.379
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr4_-_176923572
|
21.304
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr6_+_126240369
|
20.961
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_-_182357871
|
20.703
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr6_+_52529157
|
20.691
|
|
LOC730101
|
uncharacterized LOC730101
|
|
chr3_-_145968819
|
20.525
|
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr22_-_19512859
|
20.322
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr3_-_145968958
|
20.277
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr19_+_50031232
|
19.920
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr6_+_29624757
|
19.843
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr14_+_95047792
|
19.703
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr2_+_26403584
|
19.473
|
NM_001191033
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr1_+_104292439
|
19.453
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr19_-_21950322
|
19.127
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr20_-_44455932
|
19.006
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr1_+_209859524
|
18.894
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr3_+_147127151
|
18.421
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr6_+_160327972
|
17.851
|
NM_002377
|
MAS1
|
MAS1 oncogene
|
|
chr9_-_107690435
|
17.835
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chrX_+_103031433
|
17.428
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr12_+_85430098
|
17.416
|
NM_001079910
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr18_-_53303129
|
17.212
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr13_+_46626949
|
17.178
|
|
LOC100509894
|
uncharacterized LOC100509894
|
|
chrX_+_103031797
|
17.075
|
|
PLP1
|
proteolipid protein 1
|
|
chr13_+_49280619
|
17.006
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr2_+_169929009
|
16.973
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr7_-_8302141
|
16.744
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chrX_+_100805415
|
16.680
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr1_-_182357757
|
16.436
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_64920830
|
16.375
|
|
C5orf44
|
chromosome 5 open reading frame 44
|
|
chr2_+_166150340
|
16.124
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr9_+_40700290
|
16.016
|
NM_001083124
|
FAM75A3
|
family with sequence similarity 75, member A3
|
|
chr19_+_48964505
|
16.015
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr11_-_128894008
|
16.003
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_+_189839113
|
15.999
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr4_-_76944585
|
15.982
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr22_+_36113917
|
15.852
|
NM_030642
|
APOL5
|
apolipoprotein L, 5
|
|
chr5_-_147162175
|
15.799
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_+_162602198
|
15.777
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr10_+_7745229
|
15.744
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr19_+_22235278
|
15.646
|
|
ZNF257
|
zinc finger protein 257
|
|
chr1_+_1385068
|
15.510
|
NM_001039211
|
ATAD3C
|
ATPase family, AAA domain containing 3C
|
|
chr9_-_95244750
|
15.461
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr19_-_52133589
|
15.412
|
NM_003830
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5
|
|
chr2_+_189839078
|
15.284
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_+_2959511
|
14.963
|
NM_001201539
|
ARSF
|
arylsulfatase F
|
|
chr20_-_48099178
|
14.886
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr1_+_152881022
|
14.847
|
NM_005547
|
IVL
|
involucrin
|
|
chr5_+_140588267
|
14.774
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr5_-_64920100
|
14.749
|
|
TRIM23
|
tripartite motif containing 23
|
|
chr19_+_6135650
|
14.703
|
NM_030924
|
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2
|
|
chr1_-_161279724
|
14.688
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr6_+_123100645
|
14.660
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr6_-_88854989
|
14.656
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr12_-_9268513
|
14.417
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_-_15601574
|
14.364
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr3_-_58200397
|
14.315
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr8_+_22857413
|
14.149
|
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr4_+_62362838
|
14.055
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr19_+_19976625
|
14.055
|
NM_021047
|
ZNF253
|
zinc finger protein 253
|
|
chr9_+_82187469
|
14.023
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr3_-_46608039
|
14.022
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr1_+_162332772
|
13.850
|
NM_001126060
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr1_-_149889362
|
13.752
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chrY_-_27198250
|
13.749
|
NM_001002760
NM_001002761
NM_004678
|
BPY2B
BPY2C
BPY2
|
basic charge, Y-linked, 2B
basic charge, Y-linked, 2C
basic charge, Y-linked, 2
|
|
chr19_+_50030874
|
13.740
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr1_+_50569581
|
13.738
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_182922504
|
13.736
|
NM_030933
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like
|
|
chr12_+_56732658
|
13.648
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr17_+_41857802
|
13.610
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chrY_+_26764150
|
13.598
|
NM_001002760
NM_001002761
NM_004678
|
BPY2B
BPY2C
BPY2
|
basic charge, Y-linked, 2B
basic charge, Y-linked, 2C
basic charge, Y-linked, 2
|
|
chrY_+_25130409
|
13.598
|
NM_001002760
NM_001002761
NM_004678
|
BPY2B
BPY2C
BPY2
|
basic charge, Y-linked, 2B
basic charge, Y-linked, 2C
basic charge, Y-linked, 2
|
|
chr19_-_38720312
|
13.528
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr1_+_162602308
|
13.517
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr5_-_147162251
|
13.503
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr19_+_22235253
|
13.497
|
NM_033468
|
ZNF257
|
zinc finger protein 257
|
|
chr1_-_150587879
|
13.418
|
|
|
|
|
chr5_+_64920557
|
13.394
|
NM_001093755
NM_001093756
NM_001243737
NM_024941
|
C5orf44
|
chromosome 5 open reading frame 44
|
|
chr5_+_161112657
|
13.378
|
NM_000811
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6
|
|
chr12_-_10251520
|
13.347
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chrX_+_15525430
|
13.305
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chrX_-_43741593
|
13.277
|
NM_000898
|
MAOB
|
monoamine oxidase B
|
|
chr1_-_13390747
|
13.246
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr14_+_95078713
|
13.228
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chrX_-_49965657
|
13.213
|
NM_003886
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr12_-_91576805
|
13.089
|
NM_001920
|
DCN
|
decorin
|
|
chr11_-_108368918
|
13.084
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr7_+_47694841
|
12.985
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr6_-_22303014
|
12.959
|
NM_001163558
|
PRL
|
prolactin
|
|
chr12_-_10151689
|
12.937
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr4_+_55095263
|
12.823
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chrX_+_91090459
|
12.794
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr1_+_12976449
|
12.747
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr14_-_94844881
|
12.619
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_146697155
|
12.614
|
NM_001144829
NM_001144830
NM_001461
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr4_-_10041833
|
12.608
|
NM_001001290
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr12_+_72079873
|
12.535
|
NM_018279
|
TMEM19
|
transmembrane protein 19
|
|
chrX_-_65835845
|
12.488
|
NM_001242310
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr5_+_140165875
|
12.342
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chrX_-_78623006
|
12.332
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr1_+_170904611
|
12.316
|
NM_001163629
NM_025063
|
C1orf129
|
chromosome 1 open reading frame 129
|
|
chr3_+_133495925
|
12.206
|
|
TF
|
transferrin
|
|
chr7_+_142829169
|
12.084
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr21_-_39956823
|
12.083
|
NM_001243429
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_+_82186846
|
12.069
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr8_-_6837568
|
12.043
|
NM_004084
NM_001042500
|
DEFA1
DEFA1B
|
defensin, alpha 1
defensin, alpha 1B
|
|
chr5_-_151066492
|
11.974
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chrX_-_62975005
|
11.970
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr6_+_153019029
|
11.948
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr4_-_176923480
|
11.902
|
|
GPM6A
|
glycoprotein M6A
|
|
chr14_-_81687265
|
11.867
|
NM_015859
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa
|
|
chr1_-_168513201
|
11.840
|
NM_003175
|
XCL1
XCL2
|
chemokine (C motif) ligand 1
chemokine (C motif) ligand 2
|
|
chr8_-_6875777
|
11.685
|
NM_004084
NM_005217
NM_001042500
|
DEFA1
DEFA3
DEFA1B
|
defensin, alpha 1
defensin, alpha 3, neutrophil-specific
defensin, alpha 1B
|
|
chr16_-_21289571
|
11.682
|
|
CRYM
|
crystallin, mu
|
|
chr9_-_43630729
|
11.649
|
NM_001145196
|
FAM75A6
|
family with sequence similarity 75, member A6
|
|
chr1_+_104198301
|
11.591
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr9_+_134378335
|
11.518
|
|
POMT1
|
protein-O-mannosyltransferase 1
|
|
chr5_-_137475076
|
11.506
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr8_-_38008332
|
11.492
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr7_+_86273219
|
11.468
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr12_-_9268475
|
11.465
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_+_186330711
|
11.446
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr5_-_41213513
|
11.435
|
NM_000065
|
C6
|
complement component 6
|
|
chrX_+_155227245
|
11.429
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr20_+_57875481
|
11.317
|
NM_000114
NM_207032
NM_207033
NM_207034
|
EDN3
|
endothelin 3
|
|
chr3_-_187454247
|
11.316
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr22_-_27014037
|
11.300
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr1_+_205236591
|
11.194
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr8_+_133879202
|
11.171
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr8_-_17767871
|
11.135
|
|
|
|
|
chr18_-_44561967
|
11.088
|
NM_016427
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2)
|
|
chr16_+_7382750
|
11.088
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_-_75919051
|
11.047
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr5_-_64920149
|
11.028
|
NM_001656
NM_033227
NM_033228
|
TRIM23
|
tripartite motif containing 23
|
|
chr22_+_23213671
|
10.998
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr21_-_42557165
|
10.983
|
NM_182832
|
PLAC4
|
placenta-specific 4
|
|
chr3_+_44916097
|
10.918
|
NM_003241
|
TGM4
|
transglutaminase 4 (prostate)
|
|
chr18_+_61582744
|
10.891
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr10_+_57358749
|
10.791
|
NM_001190478
|
MTRNR2L5
|
MT-RNR2-like 5
|
|
chr5_-_132161863
|
10.759
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr19_-_51587466
|
10.677
|
NM_022046
|
KLK14
|
kallikrein-related peptidase 14
|
|
chr18_-_67624159
|
10.672
|
NM_006566
|
CD226
|
CD226 molecule
|
|
chrX_+_135730335
|
10.658
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chrY_+_59330251
|
10.650
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr5_-_146460991
|
10.649
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_+_15802595
|
10.637
|
NM_015849
|
CELA2B
|
chymotrypsin-like elastase family, member 2B
|
|
chr16_-_21289637
|
10.578
|
|
CRYM
|
crystallin, mu
|
|
chr14_-_50862616
|
10.566
|
NM_004196
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase)
|
|
chr5_+_156820161
|
10.514
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|