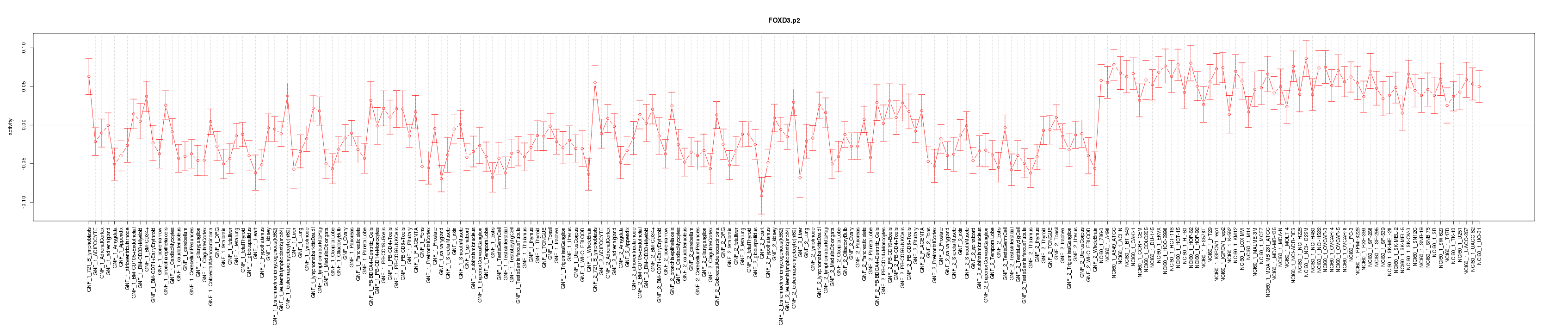
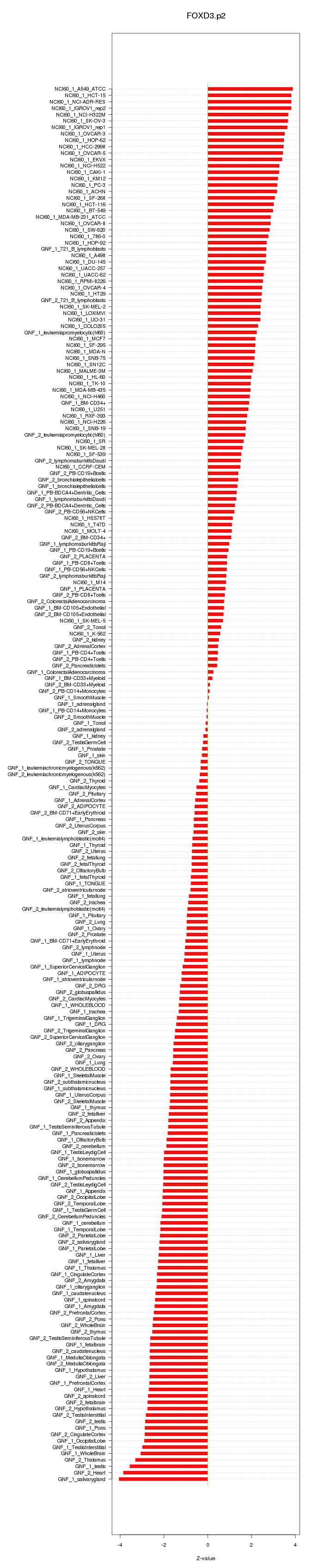
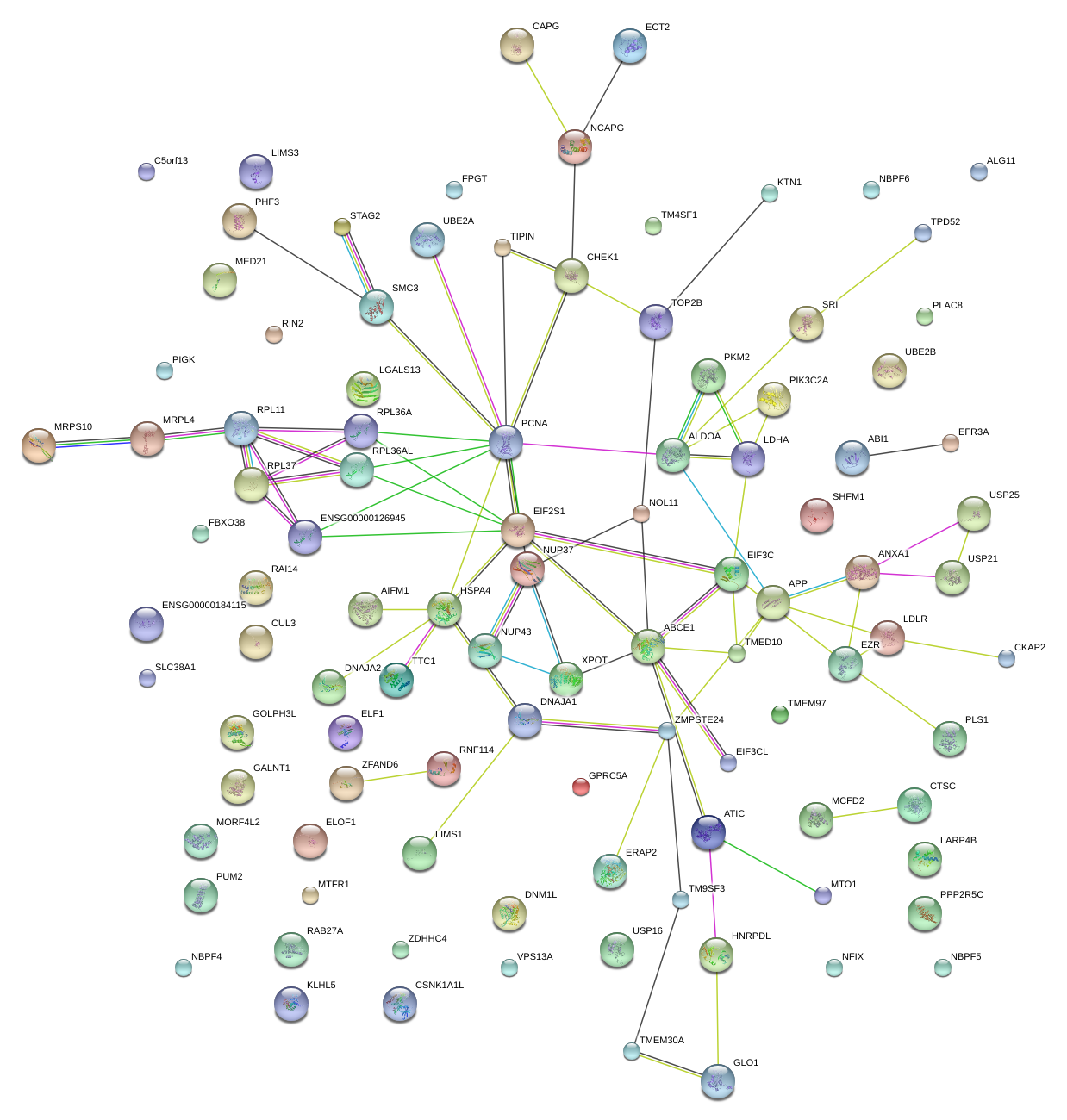
|
chrX_+_123234462
|
98.392
|
|
STAG2
|
stromal antigen 2
|
|
chr5_+_159436143
|
91.677
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr11_-_16996559
|
86.757
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr11_-_67450536
|
80.088
|
|
RPL37
|
ribosomal protein L37
|
|
chr12_+_27175482
|
61.870
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr6_+_74171610
|
60.147
|
|
MTO1
|
mitochondrial translation optimization 1 homolog (S. cerevisiae)
|
|
chr2_+_216176808
|
59.672
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr7_-_87849341
|
58.258
|
|
SRI
|
sorcin
|
|
chr4_+_146031528
|
56.972
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr2_-_47134945
|
56.556
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr14_+_102276125
|
53.752
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr3_+_172472246
|
52.583
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr2_+_216176678
|
52.046
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr5_+_159436155
|
49.728
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr2_+_216176818
|
49.619
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr6_-_150057659
|
49.419
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr22_-_43036606
|
47.743
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr3_-_25683825
|
47.395
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr16_+_30064410
|
46.386
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr20_+_19870209
|
45.230
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_+_67831484
|
44.787
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr1_-_108786663
|
44.300
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chr16_+_30064470
|
43.790
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr7_+_6617035
|
43.781
|
NM_001134387
NM_001134388
NM_001134389
NM_018106
|
ZDHHC4
|
zinc finger, DHHC-type containing 4
|
|
chr2_+_109204904
|
43.150
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_+_118714297
|
42.686
|
NM_181777
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr1_+_40735749
|
42.664
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr4_-_83347222
|
42.075
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr9_+_75766777
|
40.190
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr11_-_17191287
|
40.082
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr12_-_102512277
|
39.273
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr13_-_37679800
|
38.788
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr15_-_66648991
|
38.485
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr20_+_19915716
|
37.646
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr8_+_132991171
|
37.608
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr2_+_109204745
|
37.532
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr15_-_72501017
|
37.106
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr5_+_132406080
|
36.971
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr9_+_75766794
|
36.245
|
|
ANXA1
|
annexin A1
|
|
chr6_+_74171453
|
35.548
|
NM_001123226
NM_012123
NM_133645
|
MTO1
|
mitochondrial translation optimization 1 homolog (S. cerevisiae)
|
|
chr2_+_109204777
|
35.501
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_-_46633551
|
34.577
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chrX_-_102942968
|
34.559
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chrX_-_129272014
|
34.074
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr6_-_38670829
|
34.018
|
|
GLO1
|
glyoxalase I
|
|
chr2_-_85637486
|
33.674
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr19_+_13134782
|
33.405
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr6_-_38670882
|
32.874
|
|
GLO1
|
glyoxalase I
|
|
chr10_-_931694
|
31.658
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr14_+_102276279
|
31.477
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr12_+_64803741
|
31.330
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr2_-_20527120
|
31.172
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr6_-_75965845
|
30.814
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr16_-_28437663
|
30.795
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr11_+_18415917
|
30.692
|
NM_001135239
NM_001165415
NM_001165416
NM_005566
|
LDHA
|
lactate dehydrogenase A
|
|
chr20_+_48552928
|
29.731
|
|
RNF114
|
ring finger protein 114
|
|
chr20_+_48552902
|
29.539
|
NM_018683
|
RNF114
|
ring finger protein 114
|
|
chr20_-_5107185
|
29.050
|
NM_002592
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr16_+_28699878
|
28.988
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr14_-_75600111
|
28.800
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr14_+_56078735
|
28.679
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr21_-_27512668
|
28.613
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr11_-_88070901
|
28.204
|
|
CTSC
|
cathepsin C
|
|
chr12_+_32832209
|
28.013
|
|
DNM1L
|
dynamin 1-like
|
|
chr12_+_32832128
|
27.993
|
NM_005690
NM_012062
NM_012063
|
DNM1L
|
dynamin 1-like
|
|
chr1_-_150669499
|
27.947
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr1_+_74701070
|
27.641
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr5_+_34687663
|
27.108
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr1_-_77685076
|
26.959
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr15_+_80390757
|
26.844
|
NM_001242919
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr12_+_32832272
|
26.682
|
|
DNM1L
|
dynamin 1-like
|
|
chr11_-_88070840
|
26.608
|
|
CTSC
|
cathepsin C
|
|
chr6_-_38670904
|
26.581
|
|
GLO1
|
glyoxalase I
|
|
chr19_+_11200037
|
26.519
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr5_+_132432880
|
26.403
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr3_-_149086884
|
26.236
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr12_+_13043955
|
26.123
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr6_-_38670871
|
25.963
|
|
GLO1
|
glyoxalase I
|
|
chr18_+_33234532
|
25.903
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr16_-_47005339
|
25.599
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr10_-_98336445
|
25.565
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr10_+_112344076
|
25.386
|
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr11_+_125495030
|
25.336
|
NM_001114121
NM_001114122
NM_001244846
|
CHEK1
|
checkpoint kinase 1
|
|
chr8_+_66582107
|
25.183
|
NM_001145838
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chr5_+_147795459
|
25.048
|
|
FBXO38
|
F-box protein 38
|
|
chr12_+_27175501
|
24.935
|
|
MED21
|
mediator complex subunit 21
|
|
chr6_-_42185599
|
24.785
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr1_+_24018268
|
24.669
|
NM_000975
NM_001199802
|
RPL11
|
ribosomal protein L11
|
|
chr3_+_9476506
|
24.563
|
|
|
|
|
chr19_+_10362598
|
24.369
|
NM_015956
NM_146387
NM_146388
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr6_-_38670915
|
24.327
|
|
GLO1
|
glyoxalase I
|
|
chr13_-_41593405
|
24.086
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr13_+_52598678
|
23.873
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr20_+_48552935
|
23.758
|
|
RNF114
|
ring finger protein 114
|
|
chr13_+_53035227
|
23.677
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr13_-_41593491
|
23.573
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr5_-_111091947
|
23.538
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_42185586
|
23.438
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr17_+_65714038
|
23.096
|
NM_015462
|
NOL11
|
nucleolar protein 11
|
|
chr10_-_98325100
|
23.095
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_+_64356430
|
23.045
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr17_+_26646120
|
22.974
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr7_-_96339074
|
22.670
|
|
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1
|
|
chr2_-_86948244
|
22.493
|
NM_001198954
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough
|
|
chr10_-_27057776
|
22.437
|
|
ABI1
|
abl-interactor 1
|
|
chr8_-_80993009
|
22.379
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr11_-_88070905
|
22.105
|
NM_001114173
NM_001814
NM_148170
|
CTSC
|
cathepsin C
|
|
chr9_+_79968344
|
22.082
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr21_+_30400229
|
21.751
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr5_+_96212167
|
21.649
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr6_-_159240367
|
21.647
|
|
EZR
|
ezrin
|
|
chr4_-_84035888
|
21.596
|
|
PLAC8
|
placenta-specific 8
|
|
chr2_-_225370687
|
21.575
|
|
CUL3
|
cullin 3
|
|
chr3_+_142375738
|
21.256
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr15_-_55562483
|
20.958
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr9_-_130700762
|
20.853
|
NM_003863
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit
|
|
chr1_+_24018293
|
20.654
|
|
RPL11
|
ribosomal protein L11
|
|
chrX_+_70765479
|
20.630
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr6_-_159240433
|
20.630
|
NM_001111077
|
EZR
|
ezrin
|
|
chr11_-_64578765
|
20.576
|
NM_130803
NM_130804
|
MEN1
|
multiple endocrine neoplasia I
|
|
chr6_-_38670922
|
20.566
|
NM_006708
|
GLO1
|
glyoxalase I
|
|
chr9_+_110047049
|
20.480
|
NM_001244713
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr9_-_88715024
|
20.462
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr1_-_154578775
|
20.287
|
NM_001193495
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr3_+_133502876
|
20.116
|
NM_021203
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr3_-_105421044
|
20.084
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr2_+_109384601
|
20.056
|
|
RANBP2
|
RAN binding protein 2
|
|
chr2_-_151344145
|
20.053
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr2_-_170430312
|
20.048
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr17_-_42144986
|
20.019
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr15_+_80364902
|
19.870
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr8_-_30585442
|
19.849
|
NM_000637
NM_001195102
NM_001195103
NM_001195104
|
GSR
|
glutathione reductase
|
|
chr1_+_46016454
|
19.800
|
NM_001202413
NM_001202414
NM_006066
NM_153326
|
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase)
|
|
chr10_-_126849010
|
19.766
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr5_+_70944968
|
19.764
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr9_+_118916069
|
19.678
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr7_+_28475233
|
19.677
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_+_64680002
|
19.436
|
NM_016213
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr12_+_2904289
|
19.304
|
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr4_-_84035908
|
19.274
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chrX_+_119737905
|
19.144
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr5_+_159848835
|
19.016
|
NM_004219
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr3_+_172468469
|
19.011
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr20_-_31989303
|
19.006
|
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr6_-_42185603
|
19.003
|
NM_018141
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr2_+_172821874
|
18.768
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr2_+_201170603
|
18.357
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr15_+_57511616
|
18.302
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr4_+_41614725
|
18.288
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_77469446
|
18.241
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr4_-_139163222
|
18.229
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr15_-_60690115
|
18.188
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr6_-_159240398
|
18.160
|
|
EZR
|
ezrin
|
|
chr11_+_87032428
|
18.055
|
|
TMEM135
|
transmembrane protein 135
|
|
chr1_+_44064509
|
17.969
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr14_+_39649711
|
17.955
|
|
PNN
|
pinin, desmosome associated protein
|
|
chrX_-_23855456
|
17.860
|
|
RPL9
|
ribosomal protein L9
|
|
chr17_-_48945338
|
17.818
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr18_-_47814691
|
17.725
|
NM_001101654
NM_014593
|
CXXC1
|
CXXC finger protein 1
|
|
chr15_-_42749710
|
17.351
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr14_+_67656109
|
17.330
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr6_-_31830551
|
17.062
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr2_+_201170795
|
17.038
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr12_+_56915587
|
16.849
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr3_+_130569368
|
16.666
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr4_-_76878558
|
16.385
|
|
SDAD1
|
SDA1 domain containing 1
|
|
chr17_-_7578810
|
16.325
|
NM_001126115
NM_001126116
NM_001126117
|
TP53
|
tumor protein p53
|
|
chr20_-_31989330
|
16.208
|
NM_016082
NM_016408
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr22_-_43355886
|
16.171
|
NM_001184971
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr19_+_1026271
|
16.143
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr11_+_69467210
|
16.142
|
|
CCND1
|
cyclin D1
|
|
chr16_-_23607603
|
16.023
|
NM_005003
|
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
|
|
chr7_+_28725597
|
16.020
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr18_+_9102618
|
15.976
|
NM_021074
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa
|
|
chr1_-_161147757
|
15.905
|
NM_001199873
NM_001199874
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr17_-_30228715
|
15.769
|
NM_018428
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast)
|
|
chr9_+_127962850
|
15.684
|
NM_005833
|
RABEPK
|
Rab9 effector protein with kelch motifs
|
|
chr5_-_68665839
|
15.638
|
NM_001015891
NM_001015892
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa
|
|
chr2_-_152146373
|
15.632
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr6_-_159240399
|
15.574
|
|
EZR
|
ezrin
|
|
chr8_-_6420454
|
15.546
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr3_-_189838673
|
15.503
|
|
LEPREL1
|
leprecan-like 1
|
|
chr19_-_39826676
|
15.332
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_15839674
|
15.298
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_-_127664511
|
15.275
|
|
ECHDC1
|
enoyl CoA hydratase domain containing 1
|
|
chr2_-_74382352
|
15.265
|
|
|
|
|
chrX_+_119737743
|
15.258
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_183582667
|
15.210
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr6_-_144416633
|
15.095
|
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr12_-_80307690
|
15.093
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr2_-_10952855
|
15.093
|
NM_005742
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr2_-_100046262
|
15.045
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr4_-_170679027
|
14.999
|
NM_017867
|
C4orf27
|
chromosome 4 open reading frame 27
|
|
chr11_+_57529233
|
14.941
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr19_-_10613767
|
14.896
|
NM_203500
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr9_-_14086901
|
14.880
|
|
NFIB
|
nuclear factor I/B
|
|
chr10_-_58121011
|
14.876
|
|
ZWINT
|
ZW10 interactor
|
|
chr15_-_25650647
|
14.843
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr14_-_55658205
|
14.791
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr13_-_37573474
|
14.756
|
NM_001142364
NM_013338
|
ALG5
|
asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae)
|