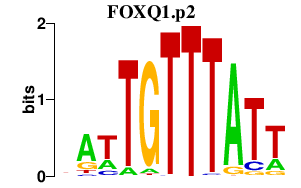
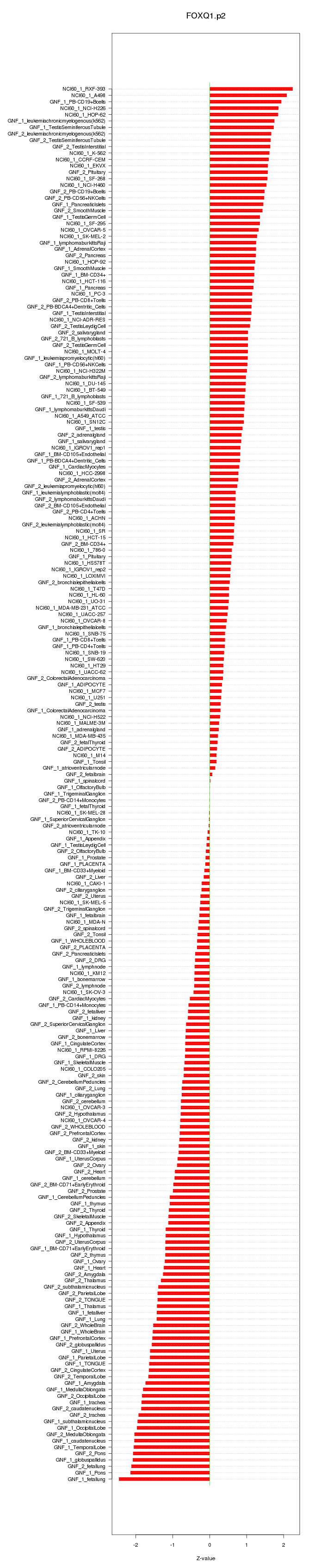
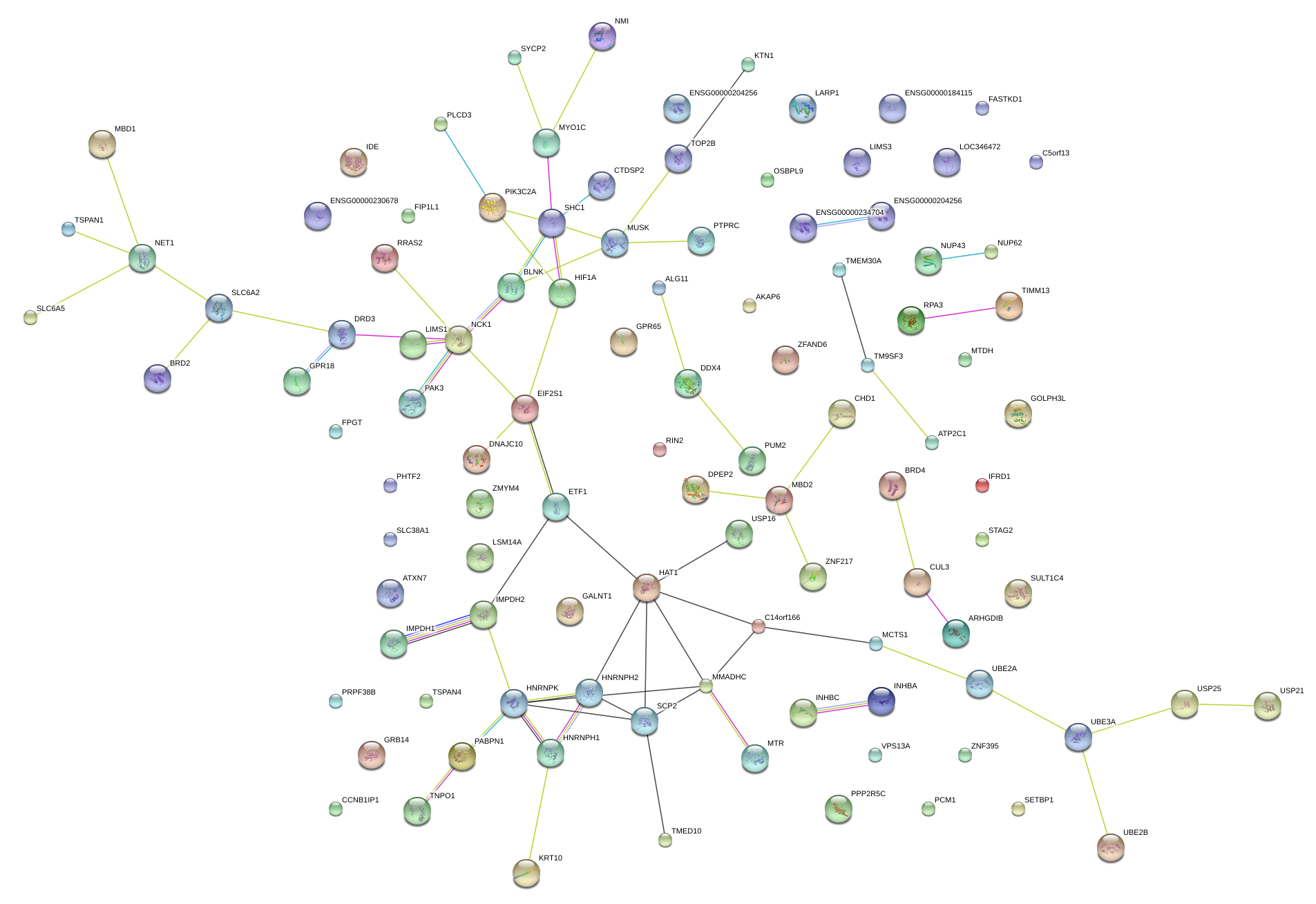
|
chr18_+_33234532
|
33.444
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr15_+_80390757
|
32.091
|
NM_001242919
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr12_-_46633551
|
23.423
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_+_52082545
|
18.212
|
NM_024586
NM_148906
NM_148908
NM_148909
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr3_-_49066858
|
16.964
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chrX_+_119738542
|
16.868
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr3_-_49066760
|
16.797
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chrX_+_123234462
|
16.691
|
|
STAG2
|
stromal antigen 2
|
|
chr14_+_67831484
|
16.410
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr5_-_137848517
|
15.954
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr11_-_17191287
|
15.454
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr3_-_49066813
|
15.422
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr2_-_152146373
|
15.258
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr9_+_79968344
|
15.211
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr5_+_72143993
|
14.754
|
|
TNPO1
|
transportin 1
|
|
chr3_+_136649316
|
14.045
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr7_-_41742666
|
14.043
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chrX_+_119737743
|
13.984
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_108994366
|
13.342
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr20_-_52199635
|
13.154
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr1_-_179833581
|
13.001
|
|
|
|
|
chr11_-_14386051
|
12.540
|
NM_001177314
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr2_+_183582667
|
12.380
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr17_-_38978824
|
12.085
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr14_+_102276279
|
12.081
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr1_+_52082755
|
12.008
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr6_-_75965845
|
11.798
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr7_+_77469446
|
11.143
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr14_+_56078735
|
11.070
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr5_+_72143929
|
10.374
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr8_+_98703361
|
10.334
|
|
MTDH
|
metadherin
|
|
chr14_+_102276125
|
10.320
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr17_+_26794838
|
10.166
|
|
|
|
|
chr20_+_19915716
|
10.136
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr13_+_52598678
|
10.075
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr14_+_52456322
|
9.905
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr10_-_98325100
|
9.856
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chrX_+_119737905
|
9.804
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_150669499
|
9.739
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr13_-_99910548
|
9.717
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr6_-_75970566
|
9.631
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr9_-_86584079
|
9.560
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chrX_+_119737984
|
9.441
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr5_-_172083569
|
9.302
|
|
|
|
|
chr1_+_35851101
|
9.255
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr10_+_5488521
|
9.120
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr2_+_109237679
|
9.068
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr14_+_88471467
|
8.951
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr7_+_112063131
|
8.942
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr5_-_98262217
|
8.891
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr12_-_15114495
|
8.819
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr1_-_154943001
|
8.701
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr10_-_98031154
|
8.593
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chrX_+_118714297
|
8.587
|
NM_181777
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr2_-_225370687
|
8.331
|
|
CUL3
|
cullin 3
|
|
chr7_-_7758237
|
8.307
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr14_-_20797532
|
8.262
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr3_+_130569368
|
8.177
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr2_-_150444219
|
8.159
|
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr2_-_150444236
|
7.995
|
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr11_+_844111
|
7.832
|
|
TSPAN4
|
tetraspanin 4
|
|
chr3_-_25683825
|
7.740
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr10_-_98290309
|
7.640
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr19_+_34710327
|
7.603
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr21_+_30400229
|
7.509
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr14_+_23789396
|
7.492
|
NM_004643
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr10_-_94257470
|
7.303
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr8_-_28243941
|
7.280
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr4_+_54292035
|
7.250
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr19_-_50432952
|
7.199
|
NM_016553
|
NUP62
|
nucleoporin 62kDa
|
|
chr12_+_57828457
|
7.181
|
NM_005538
|
INHBC
|
inhibin, beta C
|
|
chr6_-_150057659
|
7.157
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr2_-_20512016
|
7.132
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr14_-_75600111
|
7.080
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr5_-_179051586
|
7.073
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr11_+_844445
|
7.064
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr18_-_51690988
|
7.059
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr9_+_113431050
|
6.829
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr6_+_32940860
|
6.795
|
NM_001199456
|
BRD2
|
bromodomain containing 2
|
|
chr2_-_170430312
|
6.734
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr14_+_62164339
|
6.729
|
NM_001243084
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr8_+_17822093
|
6.702
|
|
PCM1
|
pericentriolar material 1
|
|
chr2_+_172821874
|
6.673
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr3_+_63953419
|
6.633
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr2_+_183608329
|
6.609
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr15_+_80352257
|
6.608
|
NM_001242913
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr1_+_74701070
|
6.559
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr5_-_111091947
|
6.536
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_154092461
|
6.395
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr1_+_236958539
|
6.337
|
NM_000254
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr2_-_165424993
|
6.202
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr15_-_25650647
|
6.152
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr17_-_73774447
|
6.109
|
|
|
|
|
chr1_+_198608136
|
6.102
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr14_+_32798478
|
6.066
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_-_225378320
|
6.047
|
|
CUL3
|
cullin 3
|
|
chr19_-_2427874
|
6.038
|
NM_012458
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr5_+_55062712
|
6.019
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr3_-_113897849
|
5.957
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr1_+_53480573
|
5.908
|
NM_001007099
NM_001007100
NM_001007250
|
SCP2
|
sterol carrier protein 2
|
|
chr6_-_87804773
|
5.855
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr18_+_21032786
|
5.849
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr13_+_29233279
|
5.849
|
|
POMP
|
proteasome maturation protein
|
|
chr12_+_64803741
|
5.787
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr12_+_50869531
|
5.768
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr1_+_53480616
|
5.756
|
|
SCP2
|
sterol carrier protein 2
|
|
chr2_-_18770776
|
5.711
|
NM_001199103
NM_001199104
NM_001002006
NM_001199086
NM_001199087
NM_001199088
NM_033253
|
NT5C1B-RDH14
NT5C1B
|
NT5C1B-RDH14 readthrough
5'-nucleotidase, cytosolic IB
|
|
chr2_-_86422591
|
5.683
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr1_+_198608244
|
5.668
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_+_13043955
|
5.661
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr4_+_68424444
|
5.591
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr12_+_14538232
|
5.534
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr4_-_140223611
|
5.461
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr1_+_154192654
|
5.410
|
NM_001127320
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr3_-_141868208
|
5.404
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr1_-_63988811
|
5.359
|
NM_001206739
NM_014288
|
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin)
|
|
chrX_+_108779009
|
5.297
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr9_+_112887780
|
5.290
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr8_+_66582107
|
5.267
|
NM_001145838
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chr10_-_91405320
|
5.250
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr5_-_83016850
|
5.241
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr15_+_89182038
|
5.227
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr20_-_34241830
|
5.189
|
NM_003915
|
CPNE1
|
copine I
|
|
chr6_+_87969619
|
5.178
|
|
ZNF292
|
zinc finger protein 292
|
|
chr2_-_200325200
|
5.133
|
|
SATB2
|
SATB homeobox 2
|
|
chr1_-_41627045
|
5.131
|
NM_001031694
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr20_+_56964162
|
5.124
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr20_+_32150139
|
5.117
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr16_-_28437663
|
5.060
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr21_-_43346780
|
5.004
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr22_+_31366662
|
4.981
|
|
|
|
|
chr4_+_169013687
|
4.952
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr7_+_102987970
|
4.905
|
NM_001204453
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr3_-_15839674
|
4.902
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr14_-_20801427
|
4.883
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr11_+_118257222
|
4.866
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr1_+_192544856
|
4.859
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr6_-_42016383
|
4.850
|
|
CCND3
|
cyclin D3
|
|
chr11_+_69467210
|
4.837
|
|
CCND1
|
cyclin D1
|
|
chr2_-_86422504
|
4.831
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr3_-_32544319
|
4.818
|
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr3_-_32544358
|
4.818
|
NM_017801
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr1_+_198608216
|
4.797
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_9913415
|
4.781
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr3_-_57234279
|
4.731
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr16_+_28699878
|
4.711
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr20_-_31989303
|
4.703
|
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr2_+_27435166
|
4.693
|
NM_001170795
NM_016085
|
C2orf28
|
chromosome 2 open reading frame 28
|
|
chr19_+_10781648
|
4.693
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr7_+_28452143
|
4.690
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_14086901
|
4.675
|
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_80307690
|
4.666
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr7_+_115863004
|
4.633
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr2_-_100046262
|
4.591
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr2_+_198380294
|
4.565
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr1_-_161147757
|
4.467
|
NM_001199873
NM_001199874
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr2_+_152214105
|
4.453
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_132406080
|
4.427
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr5_+_1801493
|
4.412
|
NM_004553
|
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase)
|
|
chr6_+_64356430
|
4.388
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr10_+_11206928
|
4.386
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_115572414
|
4.376
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr1_-_36097137
|
4.373
|
NM_001199780
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2
|
|
chr5_+_137673223
|
4.364
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr2_-_203776200
|
4.321
|
|
WDR12
|
WD repeat domain 12
|
|
chr3_+_172472246
|
4.307
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr2_-_192711651
|
4.300
|
|
SDPR
|
serum deprivation response
|
|
chr7_+_134233848
|
4.231
|
NM_001080538
|
AKR1B15
|
aldo-keto reductase family 1, member B15
|
|
chr3_+_135741576
|
4.180
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr2_+_87144737
|
4.157
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr8_-_93107643
|
4.153
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_94292737
|
4.138
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_41276112
|
4.134
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr12_-_102512277
|
4.121
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr12_+_69979268
|
4.121
|
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta)
|
|
chr6_-_131321868
|
4.115
|
NM_001252660
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr3_-_119582422
|
4.088
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr10_+_74033676
|
4.087
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr20_-_31989330
|
4.073
|
NM_016082
NM_016408
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr6_-_111888520
|
4.038
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr2_-_160472960
|
4.005
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr2_+_27440342
|
3.957
|
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr8_-_93107442
|
3.952
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_118916069
|
3.946
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr15_-_56209305
|
3.918
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr10_+_122610686
|
3.894
|
NM_018117
|
WDR11
|
WD repeat domain 11
|
|
chr8_-_145754389
|
3.854
|
NM_001001795
|
C8orf82
|
chromosome 8 open reading frame 82
|
|
chr11_+_101983170
|
3.853
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr8_-_108510094
|
3.842
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr13_-_24007822
|
3.838
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr11_-_82997376
|
3.812
|
NM_021825
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr17_-_48945338
|
3.794
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr2_+_109271267
|
3.783
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr19_-_10291056
|
3.781
|
|
|
|
|
chr3_+_187943192
|
3.771
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_-_140895407
|
3.758
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr15_-_60919642
|
3.741
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr4_-_103266577
|
3.729
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr12_-_102872329
|
3.701
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr6_+_26104103
|
3.700
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|