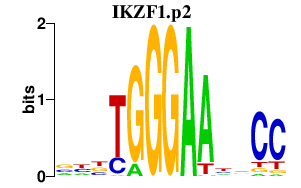
|
chr18_-_74728963
|
12.631
|
|
MBP
|
myelin basic protein
|
|
chr18_-_74729049
|
12.598
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr9_-_21994379
|
11.795
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr17_-_56618178
|
11.786
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr9_-_21994414
|
11.107
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr3_-_149688638
|
9.586
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr16_+_29690464
|
9.036
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr4_-_176733413
|
8.984
|
|
GPM6A
|
glycoprotein M6A
|
|
chr2_+_17721806
|
8.950
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr9_-_21994329
|
8.949
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr19_-_10697907
|
8.807
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr16_+_56659679
|
8.680
|
|
MT1E
|
metallothionein 1E
|
|
chr16_+_56659584
|
8.547
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr2_+_17721983
|
8.446
|
|
VSNL1
|
visinin-like 1
|
|
chr4_-_176734181
|
8.337
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_6845381
|
7.592
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr4_-_176733901
|
7.415
|
|
GPM6A
|
glycoprotein M6A
|
|
chr16_+_23690234
|
7.136
|
|
PLK1
|
polo-like kinase 1
|
|
chr8_+_145065366
|
7.101
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr15_+_41136178
|
7.014
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr2_+_47596286
|
6.932
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr9_+_130911731
|
6.931
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr16_+_29690328
|
6.881
|
NM_014298
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr4_-_120987890
|
6.680
|
NM_002358
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast)
|
|
chr12_+_48499655
|
6.571
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr12_+_52627007
|
6.470
|
|
KRT7
|
keratin 7
|
|
chr11_+_62649160
|
6.457
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr12_+_52626953
|
6.394
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr16_+_23690212
|
6.381
|
|
PLK1
|
polo-like kinase 1
|
|
chr1_+_33352071
|
6.050
|
NM_002143
|
HPCA
|
hippocalcin
|
|
chr7_-_148581359
|
6.031
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr3_+_52232174
|
5.996
|
|
ALAS1
|
aminolevulinate, delta-, synthase 1
|
|
chr3_+_52232076
|
5.973
|
NM_199166
NM_000688
|
ALAS1
|
aminolevulinate, delta-, synthase 1
|
|
chr15_-_72522062
|
5.910
|
NM_001206799
|
PKM2
|
pyruvate kinase, muscle
|
|
chr7_-_148581403
|
5.862
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr8_+_48873533
|
5.847
|
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chr19_-_42498221
|
5.798
|
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr8_-_121457625
|
5.774
|
NM_014078
|
MRPL13
|
mitochondrial ribosomal protein L13
|
|
chr6_+_34204928
|
5.755
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr21_+_41239346
|
5.701
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chrX_-_53310748
|
5.686
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr20_+_56136136
|
5.679
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr8_+_37887990
|
5.648
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr12_-_53298765
|
5.562
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr3_+_52232179
|
5.472
|
|
ALAS1
|
aminolevulinate, delta-, synthase 1
|
|
chr20_-_3149070
|
5.348
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr3_-_58563070
|
5.336
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr11_+_73357222
|
5.315
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr7_-_148581377
|
5.301
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_-_148581379
|
5.284
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr19_-_42498320
|
5.240
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr10_+_105036783
|
5.119
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr6_-_38670922
|
5.085
|
NM_006708
|
GLO1
|
glyoxalase I
|
|
chr12_-_49259549
|
5.069
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr1_-_43637788
|
5.045
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chrX_-_71792610
|
5.038
|
|
HDAC8
|
histone deacetylase 8
|
|
chr11_+_62475165
|
4.990
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr13_-_111213865
|
4.983
|
NM_017817
|
RAB20
|
RAB20, member RAS oncogene family
|
|
chr7_-_148581297
|
4.891
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_-_8931634
|
4.856
|
NM_001201483
|
ENO1
|
enolase 1, (alpha)
|
|
chr6_-_33168448
|
4.701
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr1_-_209825797
|
4.679
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr9_+_17578952
|
4.612
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr17_+_52978022
|
4.605
|
NM_005486
|
TOM1L1
|
target of myb1 (chicken)-like 1
|
|
chr1_+_155853546
|
4.572
|
|
SYT11
|
synaptotagmin XI
|
|
chr11_+_71903172
|
4.566
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr8_-_121457387
|
4.498
|
|
MRPL13
|
mitochondrial ribosomal protein L13
|
|
chr2_-_224467095
|
4.443
|
|
SCG2
|
secretogranin II
|
|
chr1_-_183274008
|
4.417
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr17_+_44068828
|
4.405
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr2_-_224467142
|
4.371
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr8_-_121457333
|
4.363
|
|
MRPL13
|
mitochondrial ribosomal protein L13
|
|
chr16_+_56703725
|
4.351
|
NM_005951
|
MT1H
|
metallothionein 1H
|
|
chr16_+_23690160
|
4.309
|
|
PLK1
|
polo-like kinase 1
|
|
chr19_+_38755097
|
4.254
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr17_-_42992852
|
4.163
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr7_-_100888367
|
4.119
|
NM_016068
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae)
|
|
chr16_+_23690191
|
4.083
|
NM_005030
|
PLK1
|
polo-like kinase 1
|
|
chr20_+_44441334
|
4.061
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr11_+_60691911
|
4.003
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr5_-_122372320
|
3.952
|
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr8_+_22224786
|
3.899
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr16_+_89989686
|
3.899
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr8_-_63951353
|
3.884
|
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase)
|
|
chr12_-_56881901
|
3.880
|
NM_013267
|
GLS2
|
glutaminase 2 (liver, mitochondrial)
|
|
chr12_-_6960406
|
3.879
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr7_-_99698916
|
3.871
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr12_+_66218141
|
3.792
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr19_+_496453
|
3.790
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr22_+_38201113
|
3.784
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr17_-_7517849
|
3.774
|
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr7_-_99698278
|
3.756
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr1_-_43637969
|
3.754
|
NM_006824
NM_001159936
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr19_+_35739282
|
3.754
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr4_-_155533850
|
3.744
|
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_143808593
|
3.744
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr8_-_63951609
|
3.673
|
NM_003878
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase)
|
|
chr7_-_100888285
|
3.653
|
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae)
|
|
chr1_-_209825673
|
3.616
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr1_+_32042085
|
3.615
|
NM_001204414
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr19_+_19322763
|
3.588
|
NM_004386
|
NCAN
|
neurocan
|
|
chr8_-_99129385
|
3.579
|
NM_005836
|
HRSP12
|
heat-responsive protein 12
|
|
chr19_+_38755422
|
3.566
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chrX_-_102941582
|
3.554
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chrX_+_47078053
|
3.548
|
NM_006201
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr4_-_155533800
|
3.544
|
|
FGG
|
fibrinogen gamma chain
|
|
chr4_-_155533882
|
3.505
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_22224758
|
3.462
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr1_+_32042122
|
3.381
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr2_-_224467049
|
3.361
|
|
SCG2
|
secretogranin II
|
|
chr6_-_42981656
|
3.353
|
|
MEA1
|
male-enhanced antigen 1
|
|
chr1_-_85930733
|
3.352
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr19_+_33182689
|
3.337
|
NM_001105570
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19
|
|
chr10_-_15413054
|
3.332
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr11_-_62475060
|
3.318
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_-_6453773
|
3.317
|
NM_007274
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr5_-_122372413
|
3.315
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr5_-_133340823
|
3.307
|
NM_003374
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr17_-_4843204
|
3.288
|
NM_001165417
NM_001165418
NM_003562
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11
|
|
chr16_-_19533285
|
3.260
|
NM_016641
|
GDE1
|
glycerophosphodiester phosphodiesterase 1
|
|
chr6_-_31514393
|
3.257
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr4_-_155533757
|
3.251
|
|
FGG
|
fibrinogen gamma chain
|
|
chr1_+_32042125
|
3.234
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr20_-_33872289
|
3.232
|
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr11_-_35440514
|
3.215
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_+_165797060
|
3.200
|
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr3_+_127317271
|
3.199
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr17_-_57784756
|
3.188
|
NM_016077
|
PTRH2
|
peptidyl-tRNA hydrolase 2
|
|
chr3_+_127317274
|
3.177
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr5_+_176875325
|
3.176
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr1_-_43637958
|
3.151
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr11_-_18270167
|
3.145
|
NM_001199744
NM_001127380
NM_030754
|
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough
serum amyloid A2
|
|
chr19_+_38755238
|
3.138
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_+_151372009
|
3.138
|
NM_002796
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4
|
|
chr1_-_85930610
|
3.134
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_-_35103144
|
3.129
|
NM_013442
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr22_+_38201232
|
3.074
|
|
H1F0
|
H1 histone family, member 0
|
|
chr20_+_44441247
|
3.000
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr1_+_153600859
|
2.990
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr17_+_38137053
|
2.990
|
NM_002809
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr5_+_36606667
|
2.990
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr17_-_1303402
|
2.980
|
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr11_+_18287807
|
2.977
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chrX_+_47077426
|
2.971
|
NM_033018
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr17_+_48712115
|
2.969
|
NM_001144070
NM_003786
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr14_+_66975205
|
2.969
|
|
GPHN
|
gephyrin
|
|
chr3_+_35722428
|
2.965
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr5_+_140186658
|
2.948
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr16_-_87903028
|
2.935
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr2_-_238499317
|
2.927
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr6_+_125474800
|
2.919
|
NM_001003396
NM_001003397
NM_003287
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr1_+_164528586
|
2.911
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr16_+_30064470
|
2.900
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr7_+_99686561
|
2.899
|
NM_006833
|
COPS6
|
COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis)
|
|
chr14_+_101295409
|
2.898
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_-_127541160
|
2.887
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr1_-_201368434
|
2.875
|
|
LAD1
|
ladinin 1
|
|
chr9_-_35103048
|
2.871
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr17_+_25799024
|
2.868
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr1_+_77747602
|
2.854
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr1_-_202612580
|
2.839
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr19_-_51869632
|
2.826
|
NM_001985
|
ETFB
|
electron-transfer-flavoprotein, beta polypeptide
|
|
chr16_+_71392615
|
2.814
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr3_-_58563490
|
2.813
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr17_+_48172508
|
2.806
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr9_-_35103089
|
2.806
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr1_+_203020310
|
2.803
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr17_+_38137115
|
2.801
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr4_-_176923480
|
2.792
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_40631846
|
2.787
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr8_+_105352050
|
2.772
|
NM_030788
|
TM7SF4
|
transmembrane 7 superfamily member 4
|
|
chr1_+_155099935
|
2.764
|
|
EFNA1
|
ephrin-A1
|
|
chr2_-_238499713
|
2.760
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr3_+_46619075
|
2.729
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr17_+_38137106
|
2.723
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr11_+_62475113
|
2.703
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr5_-_125930604
|
2.700
|
NM_001182
NM_001201377
NM_001202404
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr17_+_38137081
|
2.689
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr21_-_34960937
|
2.672
|
NM_017613
|
DONSON
|
downstream neighbor of SON
|
|
chr16_+_71392637
|
2.670
|
|
CALB2
|
calbindin 2
|
|
chr16_+_23690190
|
2.667
|
|
PLK1
|
polo-like kinase 1
|
|
chr3_+_62305395
|
2.661
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr1_+_15250578
|
2.661
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr20_+_34742656
|
2.659
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr19_-_10514242
|
2.658
|
NM_007065
|
CDC37
|
cell division cycle 37 homolog (S. cerevisiae)
|
|
chr2_+_233498206
|
2.655
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr11_+_71900958
|
2.652
|
NM_000802
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr5_-_131563481
|
2.637
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr16_-_70719854
|
2.620
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_+_150756645
|
2.609
|
NM_003040
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr11_+_66247878
|
2.603
|
NM_005700
NM_130443
|
DPP3
|
dipeptidyl-peptidase 3
|
|
chr11_+_64879294
|
2.596
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr19_-_10514154
|
2.595
|
|
CDC37
|
cell division cycle 37 homolog (S. cerevisiae)
|
|
chr4_-_56412968
|
2.581
|
NM_004898
|
CLOCK
|
clock homolog (mouse)
|
|
chr1_+_32043745
|
2.574
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr7_+_99686609
|
2.569
|
|
COPS6
|
COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis)
|
|
chr11_+_66247905
|
2.563
|
|
DPP3
|
dipeptidyl-peptidase 3
|
|
chr7_+_150756716
|
2.550
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr6_+_42981840
|
2.538
|
NM_057161
|
KLHDC3
|
kelch domain containing 3
|
|
chrX_-_72434656
|
2.526
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|